
Rice Science ›› 2016, Vol. 23 ›› Issue (6): 334-338.DOI: 10.1016/j.rsci.2016.05.004
收稿日期:2016-02-25
接受日期:2016-05-06
出版日期:2016-12-12
发布日期:2016-08-10
. [J]. Rice Science, 2016, 23(6): 334-338.
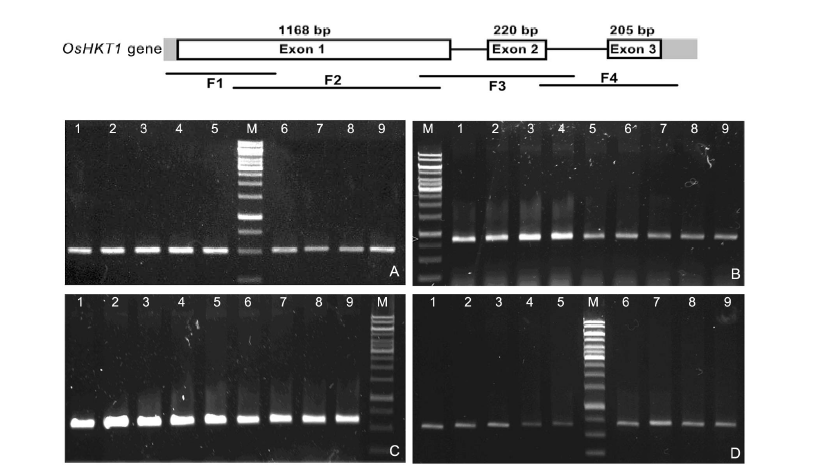
Fig. 1. PCR products using four pairs of primers. A, PCR product of 499 bp using primer pair 1 (F1); B, PCR product of 828 bp using primer pair 2 (F2); C, PCR product of 603 bp using primer pair 3 (F3); D, PCR product of 668 bp using primer pair 4 (F4). M, 1 kb ladder marker; Lanes 1 to 9, Nipponbare, Nep Non Tre, Chiem Cu, Re Nuoc, Hom Rau, Nep Oc, Ngoi, Dau An Do and Nep Deo Dang, respectively.
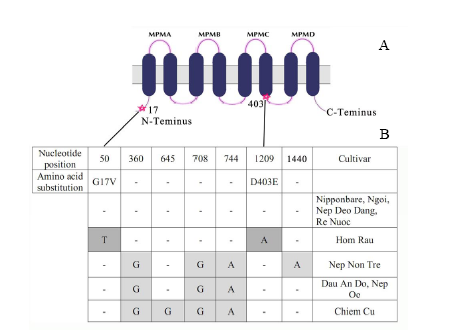
Fig. 2. Polymorphism in OsHKT1 sequence. A, Schematic representation of polymorphism locations in the protein. The position of changes in amino acid are indicated with a star; MPM, transmembrane-pore loop-transmembrane domain. B, Nucleotide sequence of OsHKT1 was compared among the cultivars with the Nipponbare sequence. Non-synonymous substitutions are indicated in dark grey, and the encoded amino acids are listed above. Synonymous substitutions are indicated in light grey. G17V means that glycine is replaced by valine; D403E means that aspartic acid is replaced by glutamic acid.
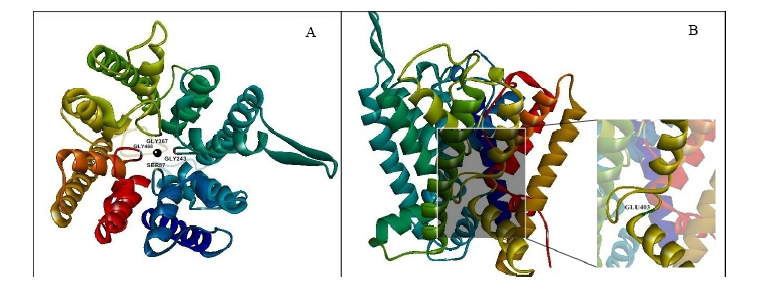
Fig. 3. 3D ribbon model of OsHKT1 protein visualized by Discovery studio 4.5 visualizer. A, 3D ribbon model of Ni-OsHKT1 protein visualized from the top that shows the pore. Four residues in Ser-Gly-Gly-Gly ion selection motif are labeled and Na+ ion is indicated as a black round circle at the center of the pore. B, Visualization of Ho-OsHKT1 transporter from the side that shows the zoom in D403E variance position (green) (G17V substitution belongs to the signal peptide so they are not modeled in 3D structure of OsHKT1 transporter).
| Position of nucleotide variance | Former codon | Usage frequency of former codon (‰) | Substituted codon | Usage frequency of substituted codon (‰) | Amino acid | Cultivar |
| 360 | CUA | 7.7 | CUG | 21.0 | Leu | Nep Non Tre, Dau An Do, Nep Oc, Chiem Cu |
| 645 | UCA | 12.4 | UCG | 12.3 | Leu | Chiem Cu |
| 708 | ACA | 11.6 | ACG | 11.4 | Thr | Nep Non Tre, Dau An Do, Nep Oc, Chiem Cu |
| 744 | ACG | 11.4 | ACA | 11.6 | Thr | Nep Non Tre, Dau An Do, Nep Oc, Chiem Cu |
| 1440 | AAG | 32.3 | CAA | 13.5 | Lys | Nep Non Tre |
| Values in bold meant that the substituted nucleotide caused an increase in codon usage frequence. | ||||||
Table 1 Changes in codon usage bias by nucleotide substitutions in nine rice cultivars
| Position of nucleotide variance | Former codon | Usage frequency of former codon (‰) | Substituted codon | Usage frequency of substituted codon (‰) | Amino acid | Cultivar |
| 360 | CUA | 7.7 | CUG | 21.0 | Leu | Nep Non Tre, Dau An Do, Nep Oc, Chiem Cu |
| 645 | UCA | 12.4 | UCG | 12.3 | Leu | Chiem Cu |
| 708 | ACA | 11.6 | ACG | 11.4 | Thr | Nep Non Tre, Dau An Do, Nep Oc, Chiem Cu |
| 744 | ACG | 11.4 | ACA | 11.6 | Thr | Nep Non Tre, Dau An Do, Nep Oc, Chiem Cu |
| 1440 | AAG | 32.3 | CAA | 13.5 | Lys | Nep Non Tre |
| Values in bold meant that the substituted nucleotide caused an increase in codon usage frequence. | ||||||
| 1 | Baxter I, Brazelton J N, Yu D, Huang Y S, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz J O, Nordborg M, Vitek O, Salt D E.2010. A coastal cline in sodium accumulation inArabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet, 6: e1001193. |
| 2 | Berthomieu P, Conejero G, Nublat A, Brackenbury W J, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, Gosti F, Simonneau T, Essah P A, Tester M, Very A A, Sentenac H, Casse F.2003. Functional analysis ofAtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J, 22(9): 2004-2014. |
| 3 | Brady K U, Kruckeberg A R, Bradshaw Jr H D.2005. Evolutionary ecology of plant adaptation to serpentine soils.Annu Rev Ecol Evol Syst, 36: 243-266. |
| 4 | Chinnusamy V, Jagendorf A, Zhu J K.2005. Understanding and improving salt tolerance in plants.Crop Sci, 45: 437-448. |
| 5 | Corpet F.1988. Multiple sequence alignment with hierarchical clustering.Nucl Acids Res, 16(22): 10881-10890. |
| 6 | Do T P, Nguyen T T T.2014. Polymorphism analysis ofOsHKT1 gene in rice(Oryza sativa). VNU J Sci: Natl Sci Technol, 30: 253-259. (in Vietnamese with English abstract) |
| 7 | Garciadeblás B, Senn M E, Banuelos M A, Rodriguez-Navarro A.2003. Sodium transport and HKT transporters: The rice model.Plant J, 34: 788-801. |
| 8 | Golldack D, Su H, Quigley F, Kamasani U R, Munoz-Garay C, Balderas E, Popova O V, Bennett J, Bohnert H J, Pantoja O.2002. Characterization of a HKT-type transporter in rice as a general alkali cation transporter.Plant J, 31(4): 529-542. |
| 9 | Hall T A.1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT.Nucl Acids Symp Ser, 41: 95-98. |
| 10 | Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A.2001. Two types of HKT transporters with different properties of Na+ and K+ transport inOryza sativa. Plant J, 27(2): 129-138. |
| 11 | Horie T, Schroeder J I.2004. Sodium transporters in plants: Diverse genes and physiological functions.Plant Physiol, 136: 2457-2462. |
| 12 | Jabnoune M, Espeout S, Mieulet D, Fizames C, Verdeil J L, Conéjéro G, Rodríguez-Navarro A, Sentenac H, Guiderdoni E, Abdelly C, Very A A.2009. Diversity in expression patterns and functional properties in the rice HKT transporter family.Plant Physiol, 150(4): 1955-1971. |
| 13 | Kelley L A, Mezulis S, Yates C M, Wass M N, Sternberg M J E.2015. The Phyre2 web portal for protein modeling, prediction and analysis.Nat Protoc, 10: 845-858. |
| 14 | Maser P, Hosoo Y, Goshima S, Horie T, Eckelman B, Yamada K, Yoshida K, Bakker E R, Shinmyo A, Oiki S, Schroeder J I, Uozumi N.2002. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants.Proc Natl Acad Sci USA, 99(9): 6428-6433. |
| 15 | Munns R.2002. Comparative physiology of salt and water stress.Plant Cell Environ, 25(2): 239-250. |
| 16 | Munns R, James R A, Laeuchli A.2006. Approaches to increasing the salt tolerance of wheat and other cereals.J Exp Bot, 57(5): 1025-1043. |
| 17 | Oomen R J F J, Benito B, Sentenac H, Rodrıguez-Navarro A, Talon M, Very A A, Domingo C.2012. HKT2;2/1, a K+-permeable transporter identified in a salt-tolerant rice cultivar through surveys of natural genetic polymorphism.Plant J, 71(5): 750-762. |
| 18 | Rains D W, Epstein E.1965. Transport of sodium in plant tissue.Science, 148: 1611. |
| 19 | Ren Z H, Gao J P, Li L G, Cai X L, Huang W, Chao D Y, Zhu M Z, Wang Z Y, Luan S, Lin H X.2005. A rice quantitative trait locus for salt tolerance encodes a sodium transporter.Nat Genet, 37: 1141-1146. |
| 20 | Rus A M, Bressan R A, Hasegawa P M.2005. Unraveling salt tolerance in crops.Nat Genet, 37: 1029-1030. |
| 21 | Schroeder J I, Ward J M, Gassmann W.1994. Perspectives on the physiology and structure of inward rectifying K+ channels in higher plants: Biophysical implications for K+ uptake.Ann Rev Biophys Biomol Struct, 23: 441-471. |
| 22 | Tarczynski M C, Jensen R G, Bohnert H J.1993. Stress protection of transgenic tobacco by production of the osmolyte mannitol.Science, 259: 508-510. |
| 23 | Tester M, Davenport R.2003. Na+ tolerance and Na+ transport in higher plants.Ann Bot, 91(5): 503-527. |
| 24 | Tran X A, Nguyen T N T, Do T P.2015. Evaluation of salt tolerance of some Vietnamese rice varieties.VNU J Sci: Natl Sci Technol, 31: 1-7. (in Vietnamese with English abstract) |
| 25 | Uozumi N, Kim E J, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker E P, Nakamura T, Schroeder J I.2000. TheArabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol, 122(4): 1249-1260. |
| 26 | Volkmar K M, Hu Y, Steppuhn H.1999. Physiological responses of plants to salinity: A review.Can J Plant Sci, 78: 19-27. |
| 27 | Yu C H, Dang Y K, Zhou Z P, Wu C, Zhao F Z, Sachs M S, Liu Y.2015. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding.Mol Cell, 59(5): 744-754. |
| 28 | (Managing Editor: Wang Caihong) |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||