
Rice Science ›› 2020, Vol. 27 ›› Issue (2): 152-161.DOI: 10.1016/j.rsci.2020.01.006
收稿日期:2019-08-05
接受日期:2019-11-13
出版日期:2020-03-28
发布日期:2019-11-28
. [J]. Rice Science, 2020, 27(2): 152-161.
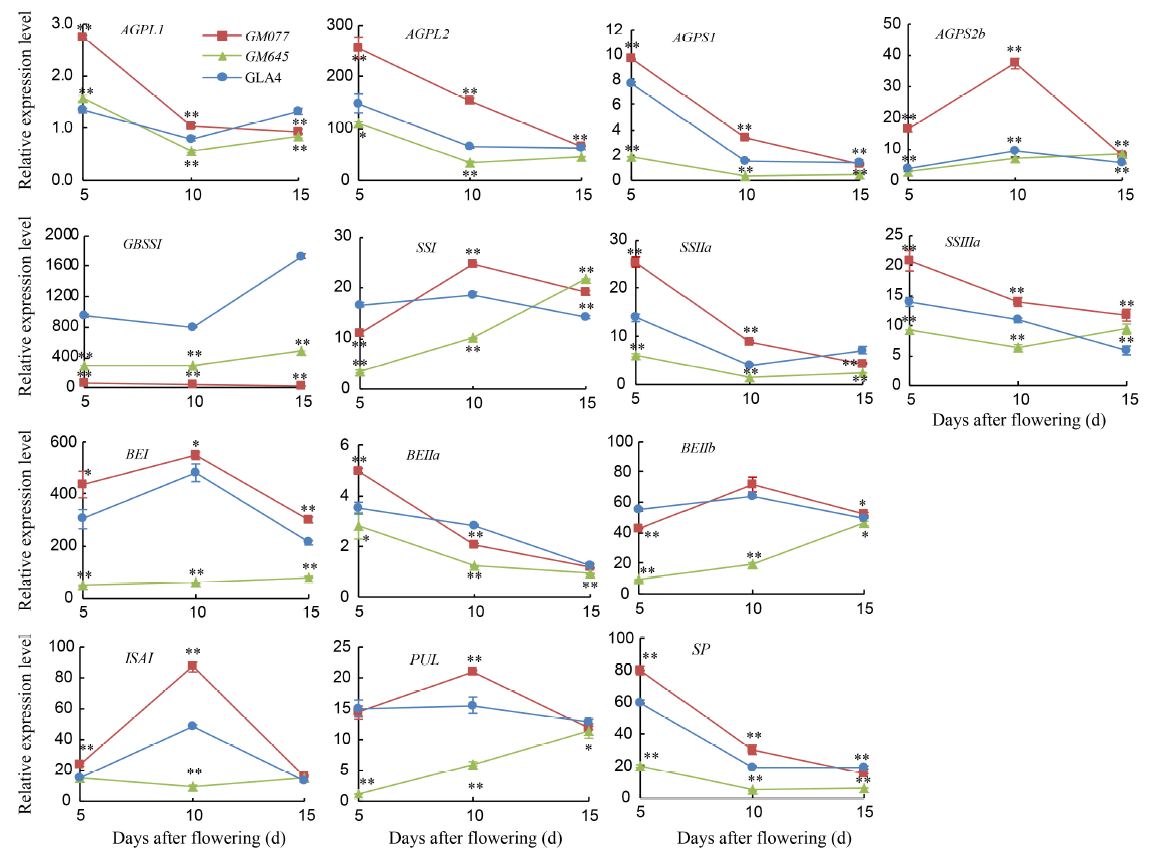
Fig. 1. Expression profiles of rice starch synthesis genes during seed development in GLA4 and two endosperm mutants. GLA4, Guangluai 4. ADP-glucose pyrophosphorylase (ADPase) genes: AGPL1, AGPL2, AGPS1 and AGPS2b; Granule-bound starch synthase (GBSS) gene: GBSSI; Soluble starch synthase (SS) genes: SSI, SSIIa and SSIIIa; Branching enzyme (BE) genes: BEI, BEIIa and BEIIb; Debranching enzyme (DBE) genes: ISAI and PUL; Starch phosphorylase (SP) gene: SP.Total RNAs were extracted from developing seeds at 5, 10 and 15 d after flowering (DAF). Data are Mean ± SD from three replicates. The asterisks indicate statistical significance between GLA4 and the mutants, as determined by the Student’s t-test (*, P < 0.05; **, P < 0.01).
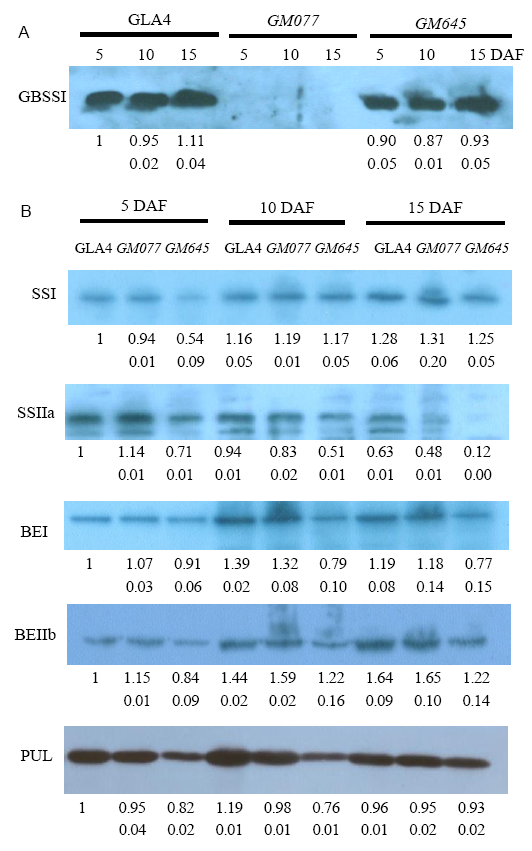
Fig. 2. Western blotting of starch synthetic enzymes extracted from developing endosperms of GLA4, GM077 and GM645 at 5, 10, 15 d after flowering (DAF) was performed using antisera against GBSSI, SSI, SSIIa, BEI, BEIIb and PUL from soluble proteins.A, Starch granule-bound protein for GBSSI blot. B, Soluble proteins for SSI, SSIIa, BEI, BEIIb and PUL enzymes blots. Values in the first line under the gel are mean relative levels to the GLA4 at 5 DAF, while those in the second lines are the standard deviation. GLA4, Guangluai 4.
| Material | Reciprocal | One sided a | No signal | ||||
|---|---|---|---|---|---|---|---|
| Strong signal | Weak signal | Strong signal | Weak signal | ||||
| GlA4 | SSI-SSIIa, SSI-BEIIb, SSI-PUL, SSIIa-BEI, SSIIa-BEIIb, BEI-BEIIb, BEI-PUL, BEIIb-PUL | SSI-BEI, PUL-SSIIa | |||||
| GM077 | SSI-BEI*, SSI-BEIIb, SSIIa-BEI, SSIIa-BEIIb, SSIIa-PUL*, BEI-BEIIb, BEI-PUL, BEIIb-PUL | SSI-SSIIa* | SSI-PUL* | ||||
| GM645 | SSI-BEIIb, SSI-PUL, BEI-PUL, BEIIb-PUL | SSI-SSIIa*, SSIIa-BEI*, SSIIa-BEIIb*, BEI-BEIIb* | SSIIa-PUL | SSI-BEI* | |||
Table 1 Comparison of protein-protein interactions among starch synthetic related enzymes in wild type and mutant endosperms determined by co-immunoprecipitation assay.
| Material | Reciprocal | One sided a | No signal | ||||
|---|---|---|---|---|---|---|---|
| Strong signal | Weak signal | Strong signal | Weak signal | ||||
| GlA4 | SSI-SSIIa, SSI-BEIIb, SSI-PUL, SSIIa-BEI, SSIIa-BEIIb, BEI-BEIIb, BEI-PUL, BEIIb-PUL | SSI-BEI, PUL-SSIIa | |||||
| GM077 | SSI-BEI*, SSI-BEIIb, SSIIa-BEI, SSIIa-BEIIb, SSIIa-PUL*, BEI-BEIIb, BEI-PUL, BEIIb-PUL | SSI-SSIIa* | SSI-PUL* | ||||
| GM645 | SSI-BEIIb, SSI-PUL, BEI-PUL, BEIIb-PUL | SSI-SSIIa*, SSIIa-BEI*, SSIIa-BEIIb*, BEI-BEIIb* | SSIIa-PUL | SSI-BEI* | |||
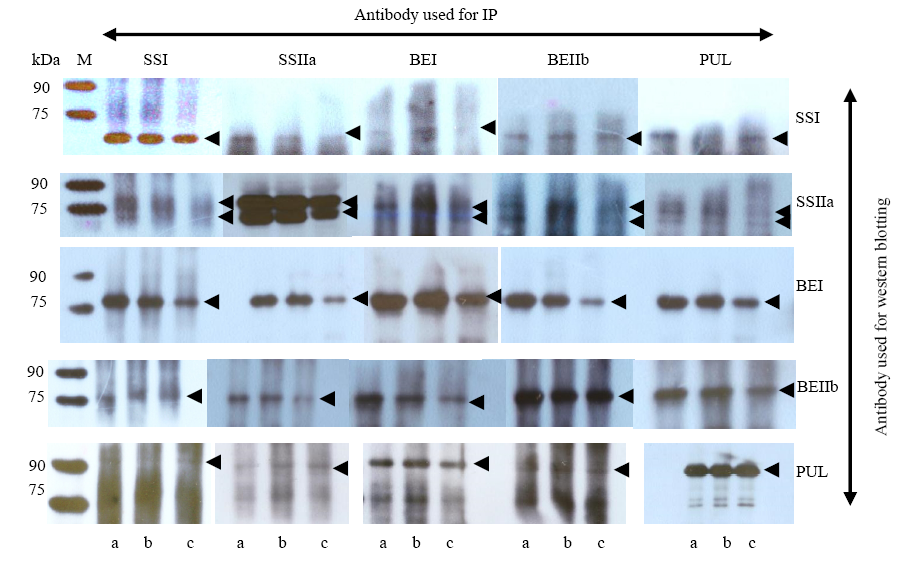
Fig. 3. Protein-protein interactions between starch synthetic related enzymes in wild type and mutant endosperms by co-immunoprecipitation (Co-IP).Black arrows represent the western blotting signals. M, Protein marker; a, Guangluai 4 (GLA4); b, GM077; c, GM645.
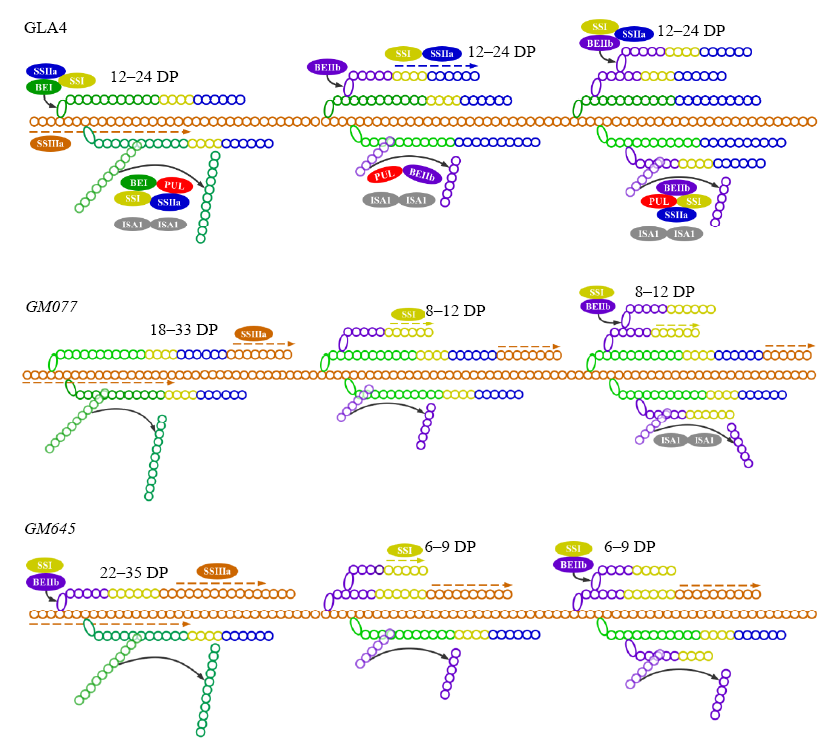
Fig. 4. Hypothesis models for starch synthesis in high amylose indica rice Guangluai 4 (GLA4) and two endosperm mutants. The predicted protein-protein complexes in developing seed of wild type indica rice GLA4 were demonstrated referring to Nakamura (2002); Nakamura et al (2005) and Fujita (2014). Only the difference in the protein-protein interactions in GM077 and GM645 from GLA4 was shown. Alterations of chain length distribution in GM077 and GM645 referred to Kong et al (2014). The small rounds of different color represent glucosyl residues from different starch synthases. DP, Degree of polymerization.
| [1] | Ahmed Z, Tetlow I J, Ahmed R, Morell M K, Emes M J. 2015. Protein-protein interactions among enzymes of starch biosynthesis in high amylose barley genotypes reveals differential roles of heteromeric enzyme complexes in the synthesis of A and B granules. Plant Sci, 233: 95-106. |
| [2] | Bao J S. 2012. Towards understanding the genetic and molecular basis of eating and cooking quality of rice. Cereal Foods World, 57(4): 148-156. |
| [3] | Bao J S. 2019. Rice starch. In: Bao J S. Rice Chemistry and Technology. 4th edn. United Kingdom and the United States: Elsevier Inc. in cooperation with AACC International: 55-108. |
| [4] | Bligh H F J, Larkin P D, Roach P S, Jones C A, Fu H, Park W D. 1998. Use of alternate splice sites in granule-bound starch synthase mRNA from low-amylose rice varieties. Plant Mol Biol, 38(3): 407-415. |
| [5] | Cai X L, Wang Z Y, Xing Y Y, Zhang J L, Hong M M. 1998. Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J, 14(4): 459-465. |
| [6] | Chen Y, Bao J S. 2016. Underlying mechanisms of zymographic diversity in starch synthase I and pullulanase in rice-developing endosperm. J Agric Food Chem, 64(9): 2030-2037. |
| [7] | Crofts N, Abe N, Oitome N F, Matsushima R, Hayashi M, Tetlow I J, Emes M J, Nakamura Y, Fujita N. 2015. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J Exp Bot, 66(15): 4469-4482. |
| [8] | Crofts N, Nakamura Y, Fujita N. 2017. Critical and speculative review of the roles of multi-protein complexes in starch biosynthesis in cereals. Plant Sci, 262: 1-8. |
| [9] | Crofts N, Iizuka Y, Abe N, Miura S, Kikuchi K, Matsushima R, Fujita N. 2018. Rice mutants lacking starch synthase I or branching enzyme IIb activity altered starch biosynthetic protein complexes. Front Plant Sci, 9: 1817. |
| [10] | Dobo M, Ayres N, Walker G, Park W D. 2010. Polymorphism in the GBSS gene affects amylose content in US and European rice germplasm. J Cereal Sci, 52(3): 450-456. |
| [11] | Du Y M, Pan T, Tian Y L, Liu S J, Liu X, Jiang L, Zhang W W, Wang Y H, Wan J M. 2019. Phenotypic analysis and gene cloning of rice floury endosperm mutant fse4. Chin J Rice Sci, 33(6): 499-512. (in Chinese with English abstract) |
| [12] | Duan M, Sun S S. 2005. Profiling the expression of genes controlling rice grain quality. Plant Mol Biol, 59(1): 165-178. |
| [13] | Fu F F, Xue H W. 2010. Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol, 154(2): 927-938. |
| [14] | Fujita N. 2014. Starch biosynthesis in rice endosperm. Agri-Biosci Monogr, 4: 1-18. |
| [15] | Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y. 2006. Function and characterization of starch synthase I using mutants in rice. Plant Physiol, 140(3): 1070-1084. |
| [16] | Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park J H, Jane J L, Hirochika H, Miyao A. 2007. Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol, 144(4): 2009-2023. |
| [17] | Fujita N, Toyosawa Y, Utsumi Y, Higuchi T, Hanashiro I, Ikegami A, Akuzawa S, Yoshida M, Mori A, Inomata K, Itoh R, Miyao A, Hirochika H, Satoh H, Nakamura Y. 2009. Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. J Exp Bot, 60: 1009-1023. |
| [18] | Hayashi M, Crofts N, Oitome N F, Fujita N. 2018. Analyses of starch biosynthetic protein complexes and starch properties from developing mutant rice seeds with minimal starch synthase activities. BMC Plant Biol, 18(1): 59. |
| [19] | Hennen-Bierwagen T A, Liu F, Marsh R S, Kim S, Gan Q, Tetlow I J, Emes M J, James M G, Myers A M. 2008. Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol, 146(4): 1892-1908. |
| [20] | Jain M, Nijhawan A, Tyagi A K, Khurana J. P. 2006. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun, 345(2): 646-651. |
| [21] | James M G, Robertson D S, Myers A M. 1995. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell, 7(4): 417-429. |
| [22] | Jeng T L, Wang C S, Tseng T H, Wu M T, Sung J M. 2009. Nucleotide polymorphisms in the waxy gene of NaN3-induced waxy rice mutants. J Cereal Sci, 49(1): 112-116. |
| [23] | Kang H G, Park S, Matsuoka M, An G. 2005. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J, 42(6): 901-911. |
| [24] | Kawasaki T, Mizuno K, Shimada H, Satoh H, Kishimoto N, Okumura S, Ichikawa N, Baba T. 1996. Coordinated regulation of the genes participating in starch biosynthesis by the rice floury-2 locus. Plant Physiol, 110(1): 89-96. |
| [25] | Kong X L, Sun X, Xu F F, Umemoto T, Chen H, Bao J S.Morphological and physicochemical properties of two starch mutants induced from a high amyloseindica rice by gamma irradiation. Starch/Stärke, 66: 157-165. |
| [26] | Kubo A, Colleoni C, Dinges J R, Lin Q, Lappe R R, Rivenbark J G, Meyer A J, Ball S G, James M G, Hennen-Bierwagen T A, Myers A M. 2010. Functions of heteromeric and homomeric isoamylase- type starch-debranching enzymes in developing maize endosperm. Plant Physiol, 153(3): 956-969. |
| [27] | Kubo A, Rahman S, Utsumi Y, Li Z, Mukai Y, Yamamoto M, Ugaki M, Harada K, Satoh H, Konik-Rose C, Morell M, Nakamura Y. 2005. Complementation of sugary-1 phenotype in rice endosperm with the wheat isoamylase1 gene supports a direct role for isoamylase1 in amylopectin biosynthesis. Plant Physiol, 137(1): 43-56. |
| [28] | Liu F, Makhmoudova A, Lee E A, Wait R, Emes M J, Tetlow I J. 2009. The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts. J Exp Bot, 60(15): 4423-4440. |
| [29] | Liu F, Ahmed Z, Lee E A, Donner E, Liu Q, Ahmed R, Morell M K, Emes M J, Tetlow I J. 2012a. Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein-protein interactions. J Exp Bot, 63(3): 1167-1183. |
| [30] | Liu F, Romanova N, Lee E A, Ahmed R, Evans M, Gilbert E P, Morell M K, Emes M J, Tetlow I J. 2012b. Glucan affinity of starch synthase IIa determines binding of starch synthase I and starch-branching enzyme IIb to starch granules. Biochem J, 448(3): 373-387. |
| [31] | Miura S, Crofts N, Saito Y, Hosaka Y, Oitome N F, Watanabe T, Kumamaru T, Fujita N. 2018. Starch synthase IIa-deficient mutant rice line produces endosperm starch with lower gelatinization temperature than japonica rice cultivars. Front Plant Sci, 9: 645. |
| [32] | Nakamura Y. 2002. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol, 43(7): 718-725. |
| [33] | Nakamura Y, Aihara S, Crofts N, Sawada T, Fujita N. 2014. In vitro studies of enzymatic properties of starch synthases and interactions between starch synthase I and starch branching enzymes from rice. Plant Sci, 224: 1-8. |
| [34] | Nakamura Y, Francisco P B, Hosaka Y, Sato A, Sawada T, Kubo A, Fujita N. 2005. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol Biol, 58(2): 213-227. |
| [35] | Nakamura Y, Utsumi Y, Sawada T, Aihara S, Utsumi C, Yoshida M, Kitamura S. 2010. Characterization of the reactions of starch branching enzymes from rice endosperm. Plant Cell Physiol, 51(5): 776-794. |
| [36] | Nishi A, Nakamura Y, Tanaka N, Satoh H. 2001. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol, 127: 459-472. |
| [37] | Ohdan T, Francisco P B, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y. 2005. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot, 56: 3229-3244. |
| [38] | Pang Y H, Zhou X, Chen Y L, Bao J S. 2018. Comparative phosphoproteomic analysis of the developing seeds in two indica rice (Oryza sativa L.) cultivars with different starch quality. J Agric Food Chem, 66(11): 3030-3037. |
| [39] | Pfister B, Lu K J, Eicke S, Feil R, Lunn J E, Streb S, Zeeman S C. 2014. Genetic evidence that chain length and branch point distributions are linked determinants of starch granule formation in Arabidopsis. Plant Physiol, 165(4): 1457-1474. |
| [40] | Ryoo N, Yu C, Park C S, Baik M Y, Park I M, Cho M H, Bhoo S H, An G, Hahn T R, Jeon J S. 2007. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep, 26(7): 1083-1095. |
| [41] | Sawada T, Itoh M, Nakamura Y. 2018. Contributions of three starch branching enzyme isozymes to the fine structure of amylopectin in rice endosperm. Front Plant Sci, 9: 1536. |
| [42] | She K C, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M, Tsuge T, Matsumoto K, Kudoh M, Itoh E, Kikuchi S, Kishimoto N, Yazaki J, Ando T, Yano M, Aoyama T, Sasaki T, Satoh H, Shimada H. 2010. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell, 22(10): 3280-3294. |
| [43] | Sun W Q, Zhou Q L, Yao Y, Qiu X J, Xie K, Yu S B. 2015. Suppressed ISA1 influences the expressions of starch synthesis related genes in the rice panicle. PLoS One, 10(3): e0122013. |
| [44] | Tetlow I J, Beisel K G, Cameron S, Makhmoudova A, Liu F, Bresolin N S, Wait R, Morell M K, Emes M J. 2008. Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol, 146(4): 1878-1891. |
| [45] | Tetlow I J, Liu F, Emes M J. 2015. Protein-protein interactions during starch biosynthesis. In: Nakamura Y. Starch: Metabolism and Structure. Switzerland: Springer: 291-313. |
| [46] | Tetlow I J, Wait R, Lu Z, Akkasaeng R, Bowsher C G, Esposito S, Kosar-Hashemi B, Morell M K, Emes M J. 2004. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell, 16(3): 694-708. |
| [47] | Toyosawa Y, Kawagoe Y, Matsushima R, Crofts N, Ogawa M, Fukuda M, Kumamaru T, Okazaki Y, Kusano M, Saito K, Ai Y, Jane J L, Nakamura Y, Fujita N. 2016. Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol, 170(3): 1255-1270. |
| [48] | Utsumi Y, Utsumi C, Sawada T, Fujita N, Nakamura Y. 2011. Functional diversity of isoamylase oligomers: The ISA1 homo- oligomer is essential for amylopectin biosynthesis in rice endosperm. Plant Physiol, 156(1): 61-77. |
| [49] | Vandeputte G E, Delcour J A. 2004. From sucrose to starch granule to starch physical behaviour: A focus on rice starch. Carbohydr Polym, 58(3): 245-266. |
| [50] | Wu Y P, Pu C H, Lin H Y, Huang H Y, Huang Y C, Hong C Y, Chang M C, Lin Y R. 2015. Three novel alleles of FLOURY ENDOSPERM2 (FLO2) confer dull grains with low amylose content in rice. Plant Sci, 233: 44-52. |
| [51] | Xu Y J, Ying Y N, Ouyang S H, Duan X L, Sun H, Jiang S K, Sun S N, Bao J S. 2018. Factors affecting sensory quality of the cooked japonica rice. Rice Sci, 25: 330-339. |
| [52] | Zhang M Z, Fang J H, Yan X, Liu J, Bao J S, Fransson G, Andersson R, Jansson C, Aman P, Sun C. 2012. Molecular insights into how a deficiency of amylose affects carbon allocation-carbohydrate and oil analyses and gene expression profiling in the seeds of a rice waxy mutant. BMC Plant Biol, 12: 230. |
| [53] | Zhong M S, Liu X, Liu F, Ren Y L, Wang Y L, Zhu J P, Teng X, Duan E C, Wang F, Zhang H, Wu M M, Hao Y Y, Zhu X P, Jing R N, Guo X P, Jiang L, Wang Y H, Wan J M. 2019. FLOURY ENDOSPERM12 encoding alanine aminotransferase 1 regulates carbon and nitrogen metabolism in rice. J Plant Biol, 62(1): 61-73. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||