Rice fields are ecosystems with many types of plants, microbes, invertebrates, birds and animals. The rice farming protects the biodiversity of the region and maintains the ecosystem for the benefit of environment. Some rice varieties release biocidal allelochemicals which might affect major weeds, microbial and pathogenic diversity around rice plants, even soil characteristics. A large number of compounds such as phenolic acids, fatty acids, indoles and terpenes have been identified in rice root exudates and decomposing rice residues, as putative allelochemicals which can interact with surrounding environment. Since these allelopathic interactions may be positive, they can be used as effective contributor for sustainable and eco-friendly agro-production system. Genetic modification of crop plants to improve their allelopathic properties and enhancement of desirable traits has been suggested. Development of crops with enhanced allelopathic traits by genetic modification should be done cautiously, keeping in view of the ecological risk assessment (non-toxic and safe for humans and ecosystem, crop productivity, ratio of benefit and cost, etc.).
In nature, microorganisms grow with multi-species communities and regulate their growth and also get influenced by them. They are rarely present as isolated species. Since ancient time, the direct and indirect chemical effects of one plant species on development of neighboring plants have been well documented. Theophrastus (300 B.C.) stated that beside reinvigorated effect of other leguminous crops on agriculture fields, Cicer arietinum have negative effect on weeds and Tribulus terrestris. Such an antipathy was also noticed between grape and cabbage plants (Culpeper, 1987). These series of observation during early time lead to the concept of allelopathy coined by Molisch (1937). According to his definition, allelopathy refers to both inhibitory and stimulatory reciprocal biochemical interactions between plants including microorganisms. However, Rice (1974) defined the term as any direct or indirect harmful effect by one plant (induding microorganisms) on another through production of chemical compounds that escape into the environment. In 1984, additional experiments and literature surveys convinced that if not all but most organic compounds that are inhibitory at some concentrations are stimulatory to the same processes at low concentrations (Rice, 1984). Further, he also differentiated allelopathy (effect depends on addition of chemical compounds in environment) from competition, which involves the removal or reduction of some factors from the environment (Rice, 1995). In 1996, the International Allelopathy Society recommended the following definition of allelopathy as: Any process involving the secondary metabolites produced by plants, microorganisms, viruses, and fungi that influence the growth and development of agricultural and biological system (excluding animals), including positive and negative effects (Torres et al, 1996).
It is now realized that complex genetic and chemical systems regulate the interactions of individual organisms within communities. The biocontrol methods based on the heavy use of synthetic chemicals have great impact on the environment. Therefore, establishing more eco-friendly methods utilizing allelopathy is one of the features for improving cultivation practices of several crops.
Inderjit and Weiner (2001) suggested that allelopathy is not just plant-plant interference but also involves soil-mediated chemical intervention. Allelopathy of soil may get influenced by many factors (physical, chemical, and biological), including the climatic conditions and presences of other plant species in the vicinity.
Rice is the most important crop worldwide, with about more than 1.5 × 108hm2 of land being cultivated for its production. Globally, rice provides approximately 20% of the caloric intake to more than 50% of the population in the world. Although rice is cultivated at such a massive scale, its yield is prone to significant loss because of infestation by weeds, pests and diseases. Out of these, yield loss because of weed infestation was reported to be more than the total loss caused by diseases and pests (Asaduzzaman et al, 2010). Therefore, in order to reduce the yield loss of rice due to weed infestation, different herbicides are reported to be incorporated in rice fields (Fig. 1; Ravi and Mohankumar, 2004). Since, extensive area is covered by rice cultivation, heavy pesticide (insecticides- 1.02 a.i/hm2, herbicide-0.19 a.i/hm2, fungicides-0.51 a.i/hm2) load enters the environment and get accumulated via leaching and biomagnifications (Shende and Bagde, 2013).
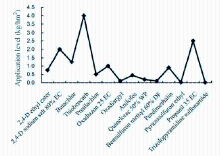 | Fig. 1. Herbicides applied for rice cultivation. Data are from Ravi and Mohankumar (2004). |
Rice fields have versatile ecotones that comprises of aquatic habitats as well as drylands and a large group of biodiversity (Fernando, 1995). In addition to the economic benefits, paddy field ecosystem helps maintain nutrient recycling, trophic structure balance, water recharge and most importantly, harbours diverse plant communities (Dhyani et al, 2007).
A few rice varieties or rice straws left in the fields after harvesting produce and release allelochemicals into the fields which suppress the growth of neighboring or successive crops/plants (Inderjit et al, 2004). Allelopathy plays a relatively better role in the competitive outcome later in the season because allelopathic interactions increase with age and density of rice. The environmental stress increases the allelopathic strength of a given plant (Waller and Einhellig, 1999). It has been observed that the amount of allelochemicals released per plant is lesser during allelopathic interaction with increased weed density (Olofsdotter, 2001a).
The objective of the present review is to discuss some physiological and molecular aspects of allelopathy which has relatively scanty information available with respect to the importance of allelochemicals in crop cultivation, particularly focusing on the possible interactions in rice field ecosystem, as well as its potential advantage in agriculture.
The suppressing effect among crops is mainly due to interference i.e. competitive and allelopathic interactions between the plant species (Sanjerehei et al, 2011). In agroecosystems, the competition for growth resources (like sunlight, soil moisture, nutrients and space) starts few days after the emergence of seedlings and becomes severe with time. The competition may be inter-species and/or intra-species. The intra-species competition generally occurs in pure crop, while inter-species competition occurs between different plant species i.e. between the component crops in mixtures/intercropping systems and/either between the crops and weeds or between the plants of the same crop sown in narrow rows or at high plant density (Narwal and Haouala, 2013).
Zero tillage in rice-wheat cropping system might have major benefits, such as improved water usage efficiency, reduced investment cost, higher yield, reduced weed population and a positive environmental effect (Mann et al, 2008). Besides, the cropping system is expanding as new crops are involved in rotations which may help to break disease and insect cycle (Hobbs et al, 2008). In production systems with no-tillage or conservation tillage, the crop residues are buried in the soil and thus the release of allelochemicals from both the growing plants and residue decomposition might act synergistically. The trend in certain regions towards no- or minimum- tillage cropping system has developed curiosity to determine the allelopathic effect of crop residues on seed germination of weeds and on production of the successive crop (Moyer and Huang, 1997). The allelochemicals released from cereal residues are reported to have inhibitory effect on seed germination of surface weeds (Jung et al, 2004).
Cover crops and mulches prevent weed growth either through allelochemicals, competition or other mechanisms that include stimulation of microbial allelochemicals, physical barriers such as obstructing light penetration and transforming soil characteristics (Hobbs et al, 2008). Cover crops have several advantages, however, if not judiciously selected and used, they can lead to significant problems in seeding of the next crop and stimulation of the pests that may ruin the following cash crop (Snapp et al, 2005). Recently, Kim et al (2013) studied the effects of winter cover crop on rice yield and total global warming potential (GWP) and suggested that cover crops with low C/N ratio, such as vetch, may be more desirable green manures to reduce total GWP per grain yield and to improve rice productivity. In rice fields, two groups of cover crops with high biomass yield, i.e. non-leguminous (Secale cerealis and Hordeum vulgare) and leguminous crops (Astragalus sinicus and Vicia villosa), are mainly used. The leguminous crops can increase the soil N content through symbiotic N fixation (Na et al, 2007), while non-leguminous ones have comparatively higher biomass productivity (Zhang et al, 2007). Some volatile allelochemicals from crucifer green manures like glucosinolates, the breakdown epithinitriles, nitriles, isothiocyanates and ionic thiocyanates have fungicidal and herbicidal activities (Vaughan and Boydston, 1997).
Intercropping is growing of two or more crops together. It has many advantages, such as higher net returns, more biodiversity, better use of resources, less probability of total crop failure and better suppressive effects on weeds, insect pests and diseases (Ali et al, 2000).
In relay cropping, the seeds of the second crops are sown before the harvest of the first crop and the second crop develops fully before the harvest of the first crop. Relay cropping in rice fields has great promise for the best utilization of residual soil moisture and improvement of soil health and nitrogen economy (Ali et al, 2014). For example, when rice cultivar Kartikshail is grown as the first crop and rabi (winter) lentil, khesari (grass pea) and mustard are grown as the second crops, weed infection of the second crops is reduced because of the allelopathic effect of Kartikshail (Kato-Noguchi and Salam, 2013).
Crop rotation has greater effects on fungi, pathogens, insects, nematodes and weed species, and therefore, it can control pests, reduce plant diseases, enhance ecosystem diversity, improve soil physical properties and crop productivity (Mamolos and Kalburtji, 2001). In crop rotation, allelochemicals produced by a preceding crop may favor or adversely affect the following crop, thus, avoiding the inhibitory effects or exploiting the favorable interactions could improve crop production (Hedge and Miller, 1990). A traditional element of crop rotation is the replacement of nitrogen through the use of green manure in sequence with cereals and other crops (Abbassi et al, 2013). If rice is planted as monocrop twice a year, it will suppress the yield of the second crop by about 25% in areas of water shortage, and rice seedlings grow poorly in a decomposed rice straw and soil mixture (Chou, 1993). Allelopathic crops when used as rotational crops, cover crops, smother crops, green manures, or mulch are helpful in reducing noxious weeds and plant pathogens and in turn improve soil quality and crop yield (Jabran and Farooq, 2013). In agroforestry, the allelopathic effects of tree species on the crop/fodder plant and crops which are planted in rotation must carefully be considered to avoid deleterious effects later (Rizvi and Rizvi, 1992).
In agriculture field, root exudation plays an important role by influencing chemical and physical properties of soil, microbial community and growth of other competitive plant species in soil. Very few reports are available concerning physiological system involved in exudation of several compounds from root cells of plants. However, some scientists suggested that plants are able to release a large range of compounds through plasmalemma or endoplasmic derived exudation and proton-pumping mechanisms (Bais et al, 2004).
Rice variety and origin may regulate allelopathic behavior, for example, japonica rice is more allelopathic than indica and japonica-indica hybrids (Khanh et al, 2007). Rice can produce and secrete different allelochemicals into its neighboring environment with various biological effects. Therefore, it would be a vital step to understand several interactions persisting in rice field in order to utilize allelopathic properties of rice in a direction to improve its cultivation and harvest.
Many crops have been reported to be allelopathic towards other crops grown either simultaneously or subsequently (Khanh et al, 2005). Therefore, many crops have been examined specifically, for allelopathic activity towards weeds or other crops. A variety of allelochemicals released by a range of temperate and tropic crops i.e. alfalfa (Medicago sativa), barley (Hordeum vulgare), clovers (Trifoliumspp., Melilotusspp.), oats (Avena sativa), pearl millet (Pennisetum glaucum), rice (Oryza sativa), rye (Secale cereale), sorghums (Sorghumspp.), sunflower (Helianthus annuus), sweet potato (Ipomoea batatas) and wheat (Triticum aestivum), have been reported to produce suppressive effects on weeds (Dilday et al, 1994; Weston, 1996). Some crops produce allelopathic compounds when they are growing or decomposing and inhibit the growth of neighboring plants.
Various allelochemicals released by crops are considered to be environmentally safe and provide unique molecular targets as compared to synthetic pesticides. Among allelopathic crops, crucifers are considered to be the most important since the glucosinolates found in crucifers, which on hydrolysis release isothiocyanates, are a promising inhibitor of mycelial growth of Botrytis cinerea, Rhizopus stolonifer, Monilinia laxa, Mucor piriformis and Penicillium expansum, all fungal pathogens of fruit and vegetable crops (Mari et al, 1993). Similarly, Brassica is known for high concentration of allyl-isothiocyanate which restricts the growth of Fusarium sambucinum, a fungus causing potato tuber dry rot (Mayton et al, 1996). Besides this, leachates of sunflower have the potential to suppress germination and growth of Parthenium hysterophorus, a noxious weed (Kohli, 1993).
The inhibitory action of several rice varieties was recorded on different plant species, both in fields and laboratory experiments (Dilday et al, 1998; Olofsdotter et al, 1999). In paddy fields, intercropping of semi- aquatic crops like lettuce is a normal practice. With respect to such agricultural practice, it is very important to study the allelopathic potential of rice cultivars since root exudates of rice cultivars contain allelochemicals affecting germination of lettuce (Ma et al, 2014). Water soluble allelochemicals of Oryza glumaepatulahusk stimulate shoot growth of Eclipta thermalis, but inhibit the root growth of Lactuca sativa (Hiroshi, 2008). The influence of allelochemicals from 15 rice varieties was examined on the growth of spinach with the highest allelopathic potential for the rice cultivar WITA12 (Kabir et al, 2010). Different concentrations of rice extract in water show different rates of inhibition on root growth of radish, wheat and lettuce seedlings (Tarek, 2010). In order to reduce the detrimental impact of sunflower residue, allelopathic tolerant rice and wheat should be cultivated (Bashir et al, 2012). Despite the fact that rice root exudates have a large inhibitory effect on wheat seedling growth, even at the concentration of 1 mg/L (Olofsdotter, 2001b; Ma et al, 2014), rice-wheat rotation is a common agriculture practice followed all over the world (Hobbs et al, 2008). Time of sowing, crop rotation, cover-crop management, intercropping, no-till planting and nonrotational cropping systems are also involved with allelopathic effects (Khanh et al, 2005).
Generally, the farmers keep the rice residue stay in the soil after harvest since they believe the residue will benefit the growth of the subsequent crops by improving the nitrogen content and organic matter of soil. However, this practice has led to suppression of several subsequent crops since decomposition of rice straw is known to release phytotoxic chemicals (Inderjit and Dakshini, 1999). In continuous monocropping of rice, the yield has been found to significantly lower in the 2nd year than in the 1st year. Probably this could be due to autotoxic mechanism induced by decomposition of rice straw that is left in the agricultural soil after harvest (Chou, 1995). It is well documented that the toxic compounds, like p-coumaric, p-hydroxy benzoic, syringic, vanillic, ferulic and o-hydroxy phenyl acetic acid, are released directly or indirectly during microbial decomposition of rice residues (Chou and Lin, 1976). The toxicity of these compounds persists for 16 weeks after decomposition (Chou et al, 1977) and can inhibit the growth of rice seedlings (Yang et al, 2002), leaf expansion (Blum and Rebbeck, 1989), root elongation (Pramanik et al, 2000), and interact with nutrients mainly nitrogen (Chou et al, 1982). The soil in paddy fields is deficient in oxygen due to water logged condition and presence of decomposing rice residue, which creates negative redox potential in the soil. This negative rice-rice residue interaction hinders root growth of rice, however, the rice plants have adopted a survival strategy to capture required oxygen through swelling of root cells (Chou, 1995).
Chou and Lin (1976) asserted that aqueous extracts of decomposing rice residues in soil inhibit root growth of lettuce and rice seedlings. Similarly, Kayode and Ayeni (2009) conducted laboratory studies by using extracts of crop residues (sorghum stem and rice husks) to check its allelopathic effect on growth of maize (Zea mays L.). The results revealed that the seed germination, including the growth of the radical and plumule, is retarded in the extract-treated seeds as compared to the control.
Soil is a dynamic system, and one mechanism of interference is unlikely to explain plant interference in nature. Therefore, in order to understand plant interference, it is important to recognize synergistic action of several mechanisms, such as soil interference, interference allelopathy, resource competition and nutrient demineralization (Inderjit and del Moral, 1997). Apart from the direct toxic effect on other plants, some allelochemicals are supposed to influence the availability of nutrients in the soil. It is possible that the effect of allelopathic plants can be due to the allelochemicals in the soil and/or to altered soil nutrients.
Soil provides nutrition to plants and contains microorganisms, such as bacteria, fungi, algae and nematodes, which interact to facilitate nutrient acquisition (Richardson et al, 2009). Mineral nutrients are present in the soil in various forms and solubility. Under nutrient-limited environments, plants interact with soil microorganisms and release allelochemicals, which facilitate nutrient solubilization (Jones and Darrah, 1994). Phenolics are an important group of root-exuded allelochemicals (D’ Arcy-Lameta, 1986), which trigger solubilization and release of Fe, P and other nutrients, thus helping the plants to improve uptake of respective nutrients.
Crop allelopathy may be largely influenced by composition and concentration of allelochemicals, which are released from crops and degraded by several soil factors (biotic and abiotic) (Cheng, 1995). Transport, transformation, retention in rhizosphere, leaching and residual effects of rice allelochemicals are influenced by soil texture (Inderjit and Dakshini, 1994), temperature (Chou et al, 1991) and different chemicals (Cheng, 1995).
The presence of allelochemicals in a plant and its rhizosphere is not strong evidence for direct plant- plant allelopathy, because the observed growth pattern may be due to the influence of these compounds on soil ecological processes rather than direct effects on the target plants. Phenolic acids, p-coumaric, ferulic, p-hydroxybenzoic and protocatechuic acids influence the accumulation of soil organic N and inorganic ions such as Al3+, Fe2+, Mn2+ and PO43- (Inderjit and Mallik, 1997).
Allelopathic rice varieties produce certain rhizospheric allelochemicals in soil which can control the growth of barnyard grass and respond to some allelochemicals excreted by barnyard grass. This was evidenced by Kong et al (2006), which did not show any stimulation of such allelochemicals in non-allelopathic rice varieties.
Rice (1984) suggested that during rotation, the rate of nitrification is reduced, perhaps due to the presence of allelochemicals that subsequently depress the rates of nitrate accumulation or reduce the nitrifier populations. However, an increase in nitrification regulated by available ammonium has been reported during initial stages of rotation (Robertson and Vitousek, 1981). Allelopathy offers an attractive and natural option to decrease nitrification for improving nitrogen use efficiency in agriculture systems. Incorporating residue of various crops into soil and release of allelochemicals from plant roots may help suppress nitrification process, by inhibiting the activities of vital enzymes such as ammonium momo-oxygenase and hydroxylamine oxidoreductase (Subbarao et al, 2009) and the phenomenon known as biological nitrification inhibition (BNI) (Subbarao et al, 2006). However, in addition to root exudates, plant water extracts can also suppress the process of nitrification in soil (Alsaadawi, 2001). Allelochemicals, such as methyl 3-(4-hydroxyphenyl) propionate (Zakir et al, 2008), linoleic acid, α -linolenic acid, methyl-p- coumarate and methyl ferulate, are responsible for BNI (Subbarao et al, 2009). Phenolics and terpenoids may play an important role in the inhibition of nitrification (White, 1994). Phenolic compounds, such as caffeic and ferulic acids, myricetin, tannins and tannin derivatives, inhibit the oxidation of NH4+to NO2- by Nitrosomonas (Rice, 1984). Alternatively, it has been proposed that terpenoids enhance immobilization of ammonium-N by soil organisms rather than by inhibition of nitrification (Bremner and McCarty, 1988). Therefore, allelopathic potential of rice can be exploited to improve nitrogen use efficiency of soil.
Phytotoxicity of phenolic acids is influenced by different factors including soil type, soil pH and mineral nutrition, and other resources present in the substrate (Blum, 1998). Soil-plant debris bioassays are often employed to demonstrate phytotoxicity and allelopathy (Blum, 1999), and there are many reports of inhibitory effects of the secondary compounds released by plant debris. Plant waste materials may influence nutrient mobilization and soil pH, which can further affect nutrient immobilization and microbial activity (Aarino and Martikainen, 1994). Chemicals released by plants may influence microbial ecology through their effects on soil microbes and plant pathogens (Einhellig, 1996). Population densities of soil-borne microorganisms are affected by the soil enrichment with different phenolic acids-ferulic, p-coumaric, p-hydroxybenzoic and vanillic acids (Blum and Shafer, 1988). However, the effect was dependent on the concentrations of phenolic acids and inorganic ions in the soil.
Microorganisms play a vital role in allelopathic interactions as they can easily alter or transform the released allelochemicals through their metabolic processes (Pellissier and Souto, 1999). For example, phenolic acids have been found to be transformed by microbes via addition or deletion of side groups and polymerization. In this process of transformation (metabolism of phenolic acids and/or incorporation of carbon from other phenolic acids into microbial biomass), other organic molecules are generated which may differ in their phytotoxicity (Blum et al, 1999).
Roots also secrete allelochemicals in the soil at a significant level to interact with microorganisms and modify the microbial community of the soil (Farrar et al, 2003). In soil-system, specific microflora monitors the degradation of a particular allelochemical, while some microbial species may take advantage of allelochemicals present in the soil (Kong, 2008).
Bacteria such as Streptomyces sagononensis, S. hygroscopicum and Pseudomonas fluorescences are allelopathic and may inhibit the growth of plants present in their ecosystem. The allelochemicals from microorganisms are generally nonspecific on the growth of several annual and perennial species (Hoagland, 1990). They may be effective at a very low concentration but have variable effects on different cultivars (Ambika, 2013).
There are many different classes of microorganisms i.e. cyanobacteria and algae (Nostoc sp., Anabaena sp., Pandorina sp. and Caulerpa sp.), protozoans (Entamoeba hystolytica), viruses (Cyanophages), fungi (Epidermophytonsp., Microsporumsp. and Trichophyton sp.) and bacteria (Desulphuvibrio sp., Beggiatoa sp., Clostridium sp. and Pseudomonas denitrificans) in paddy fields (Oyewole, 2012). Rice allelochemicals have been widely studied in relation to their effects on the growth of weeds (Chung et al, 2006), but their fate and impact on microorganisms, which play an essential role in rice field ecosystem, remain obscure. Bai et al (2000) reported that dynamics of microorganisms in paddy soils greatly varies with various traits, growing periods and seasons. Organic compounds produced from rice roots monitor the microbial biomass and population in rice soils (Lu et al, 2002). Likewise, the allelochemicals released through roots of allelopathic rice could provide carbon to interact with the soil microorganisms. Rice allelochemicals would likely have a great impact on soil microorganisms once released. However, rice- microbe interactions mediated by allelochemicals in paddy soils have not yet been clearly identified and understood (Kong, 2008).
The blue-green algae (cyanobacteria) are of much ecological importance in paddy fields, maintaining soil fertility through nitrogen fixation (Nirmal Kumar et al, 2010) and even reclaiming alkaline soils (Singh, 1961). Rice straw residue has been known to release some allelochemicals during their decomposition that affect the growth and nitrogen fixing potential of blue-green algae (Rice, 1984; Ahluwalia and Ghawana, 1998). Increased organic matter and decreased pH of the soil, as well as the incorporation of paddy straw, support the growth of green algae rather than cyanobacteria (Karaush, 1985).
Decomposition of rice residue and straw in soils may produce different phenolic compounds that can have synergistic suppressive effect on Rhizobium strains by reducing their nitrogen fixing ability (Jabran and Farooq, 2013). The basal portion of rice plants left in the fields due to use of harvester combines is ploughed back or burnt. These residual part of rice plants on decomposition release water soluble phenolic compounds that might affect the algal dynamics and the germination of the next crop (Ahluwalia, 1998, 2013). Kong et al (2008a) found that 5, 7-4′ -trihydroxy-3′ , 5′ -dimethoxyflavone can control the microbial population and community structure in rice soil. It also suggests that flavone can reduce microorganisms especially fungi presented in paddy soil, whereas benzoic acid can induce a higher response for soil microorganisms especially for bacteria.
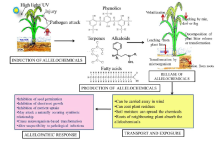
Allelochemicals present commonly by a conjugated form in almost all plants and any part of the plants, like leaves, culm, flowers, fruits, seeds, buds, pollen and roots (Putnam, 1988). Plants or organisms respond to different stimuli through synthesis and release of the allelochemicals. These chemicals are released into the environment by means of volatilization, foliar leaching, root exudation, decomposition of plant residue and debris incorporation into soils (Chou, 1990). The induction, release, transport and exposure of allelochemicals and their putative effects has been summarized in Fig. 2. These can lengthen survival times in a hostile environment and serve as defensive weapons to prevent damage and decay of reproductive organs. These can be hydrophilic or hydrophobic, absorbed to soil surface, coat plant residues, and carried away in the wind. Once released, the allelochemicals diffuse into the soil and are transported by water. These are also found in the rhizosphere soil and have been demonstrated to show allelopathic interactions between organisms through root to root contact (Inderjit and Weston, 2003).
Allelochemicals are largely classified as the secondary plant metabolites which are generally considered as alkaloids, phenolics, flavinoids, terpenoids and glucosinolates. These do not play a role in the primary metabolic process of the plant, but are essential for their survival (Rice, 1984).
Several allelochemicals like 5-hydroxy-2- indolecarboxylic acid, 5-hydroxyindole-3-acetic acid, mercaptoacetic acid, 4-vinylphenol, trans-ferulic acid (Song et al, 2004), ergosterol peroxide, 7-oxo- stigmasterol (Macias et al, 2005), 5, 7-4′ -trihydroxy- 3′ , 5′ -dimethoxyflavone (Kong et al, 2006), 3-hydroxy- β -ionone, 9-hydroxy-4-megastigmen-3-one (Kato- Noguchi et al, 2011) azelaic acid, tetradecanoic acid, 1, 2-benzenedicarboxylic acid bis (2-ethylhexyl) ester, 1H-indole-3-carboxylic acid, 1H-indole-5-carboxylic acid, 1H-indole-3-carboxyaldehyde, 3, 4-dihydro- xyhydrocinnamic acid, 3-hydroxy-4-methoxybenzoic acid, 4-hydroxycinnamic acid and 4-hydroxy- hydrocinnamic acid (Rimando et al, 2001) have been reported in rice. Number of reports on rice allelochemicals indicate their nature as phenolic compounds and momilactones. It has been accepted that the role of momilactones (Kato-Noguchi and Ino, 2005) with phenolic compounds (Seal et al, 2004a) and some unknown compounds in rice plants may be responsible for their allelopathic activity (Table 1).
| Table 1 Production of various allelochemicals from rice (Oryza sativa). |
Seal et al (2004a) reported 200 different compounds in rice root exudates and put these under the three main chemical classes (phenolics, phenylalkanoic acids and indoles), and several classes of the secondary metabolites determined from the root exudates like phenolics (Rimando et al, 2001; Inderjit et al, 2002), alkyl resocinols (Bouillant et al, 1994), momilactone B (Kato-Noguchi and Ino, 2005), carbohydrates and amino acids (Bacilio-Jimenez et al, 2003) and flavones (Kong et al, 2004a). Flavones and their O-or C- glycosides, cyclohexenone, and momilactones play a key role in rice allelopathy (Kong et al, 2004a, b) by generating many biological and ecological effects (Chung et al, 2005). It has been found that the flavones play a role in species interaction between rice and other organisms, particularly in the rhizosphere and have the highest inhibitory action as compared to phenolics (Bais et al, 2006). Among the other allele- chemicals involved in rice allelopathy, 5, 4′ -dihydroxy- 3′ , 5′ -dimethoxy-7-O-β -glucopyranosylflavone is unique because a significant amount could be exuded from the roots to the rihzosphere and then rapidly transformed into aglycone (5, 7-4′ -trihydroxy-3′ , 5′ - dimethoxyflavone), which will ultimately produce many biological effects by interacting with soil organisms (Kong et al, 2008a, b). Bran-containing brown rice produce tricin (5, 7-4′ -trihydroxy-3′ , 5′ - dimethoxyflavone), an allelochemical suspected to possess cancer chemopreventive properties (Cai et al, 2005). Rice seedling rot disease is a dominant problem in rice cultivation due to frequent use of direct seeding practice. The tricin produced by rice hulls were found to have a fungicidal effect on Fusarium oxysporumand Rhizoctonia solani(soil-borne pathogenic fungi causing rice seedling rot disease). However, this effect was higher with aurone, an isomer of tricin as compared with tricin itself (Kong et al, 2010).
Momilactone B was found in root exudes of allelopathic rice cultivars, PI312777, with 5, 7-4′ - trihydroxy-3′ , 5′ -dimethoxyflavone and 3-isopropyl- 5-acetoxycyclohexene-2-one-1 (Kong et al, 2004a). In addition, momilactone A, another potential allelochemical, was found in rice root exudates of Oryza sativa cv. Koshihikari (Kato-Noguchi et al, 2008b). Momilactones A and B were first isolated from rice husks as growth inhibitors (Takahashi et al, 1976), but later, also found in rice leaves and straw as phytoalexins (Lee et al, 1999).
Several studies focused on the common putative allelochemicals found in rice like phenolic acid compounds (p-coumaric acid, p-hydrobenzoic acid, feruic acid and vanillic acid) (Rimando et al, 2001; Seal et al, 2004b). However, concentration of single phenolic acid and combination of several phenolic acids measured in rice ecosystem do not approach phytotoxic levels (Olofsdotter, 2001a). Furthermore, it was recorded that allelopathic rice cultivars can’ t release significant amount of penolic acids than non-allelopathic cultivars (Olofsdotter et al, 2002a). The target sites of allelochemicals play an important role in breeding process since it will help to decide which chemical to increase in order to achieve the desired output. The action mechanism of some rice allelochemicals are described in Table 2.
| Table 2 Mode of action of major rice allelochemicals. |
Weeds pose an important biological constraint to crop productivity. Many weeds release allelochemicals to interfere with crop plants. These allelopathic weeds are economically destructive, and the attempt to control them has met with limited success. However, the allelopathic action may be used as an important strategy for crop and weed management system. Weeds cause reductions in yield and quality and remain one of the biggest problems in rice production. The negative impact of commercial herbicides makes it desirable to search for other alternative weed management options (Nirmal Kumar et al, 2010), and allelopathy seems to be one of the options (Tesio and Ferrero, 2010). Momilactone B inhibits the growth of typical rice weeds like Echinochloa crus-galliand E. colonumat concentrations greater than 1 μ mol/L (Kato-Noguchi et al, 2008a).
Improved allelopathic potential of a crop may be useful for rice and other crops (Olofsdotter et al, 1999). If crop allelopathy is applied in crop rotation system, it can prove to be a successful tool to manage weed infestation in agricultural production (Khanh et al, 2005). Despite of this, crop rotation system in paddy field is difficult since most crops may not survive in moist soil (Tuten, 2010). Therefore, exploiting rice itself through isolation and identification of allelochemicals, responsible for weed suppression, will be the most feasible means of weed control (Khanh et al, 2013).
Paddy weeds such as barnyard grass (Echinochloa crus-galli), oval-leafed pondweed (Monochoria vaginalis), redstem (Erodium cicutarium) and ducksalad (Heteranthera limosa) are the most common species used as indicator plants, as they reflect an actual rice-weed interaction. Utilization of rice residues in paddy fields has long been recognized as an important source to improve the status of organic matter of soil and was also reported to reduce the emergence of weeds. To date, decomposition of rice straw and stubble has reduced the occurrence of both broadleaved and grassy weeds (Narwal, 2000). Incorporating the residues of rice with high allelopathic activity, minimised rice flatsedge (Cyperus iria L.) growth to a similar degree than that achieved by the application of propanil and bentazon herbicides (Lin et al, 1992). Leaves plus straw and hulls of some rice cultivars with strong allelopathic property dramatically inhibit weed interference by about 60%-95% (Jung et al, 2004). Furthermore, another trial showed rice residues (variety Sarjoo 52) blended into the soils (5-6 cm in depth) suppress jungle rice (Echinochloa colona (L.) Link), monarch redstem (Ammania baccifera L.), many flowered ammannia (A. multiflora Roxb.) and gulf leaf flower (Phyllanthus fraternus Webster) (Khan and Vaishya, 1992). Straw, leaves and hulls of some rice cultivars suppress the germination of field bind weed (Convolvulus arvensis) and littleseed canary grass (Phalaris minor) (Ahn and Chung, 2000; Inderjit et al, 2004). The use of allelopathic rice cultivars and allelochemicals can definitely reduce the ecological impact, particularly by reducing the amount of herbicide to be used. In most allelopathic rice cultivars, more than one allelochemical is available and may play a role for inhibition of weeds.
Pheng et al (2010) conducted pot experiments to evaluate the response of the rice line ST-3 and three weed species, barnyard grass (E. crus-galli), small umbrella sedge (Cyperus difformis), and water primrose (Ludwigia octovalves), to the residues of 16 rice lines. Later, the field studies were performed in order to determine the response of the same rice line and weed species to the residue of seven putative allelopathic rice lines and one non-allelopathic rice line. It was observed that if the residue’ s incorporation was delayed by two weeks or only a proportion of the residue was incorporated, the rice crop could withstand the growth-inhibiting effect, while the inhibition of the weed species was retained.
Allelopathic rice cultivars combined with cultural management options are therefore, interesting and potential strategy contributing to alternative chemical control of weeds in paddy ecosystems (Olofsdotter et al, 2002b). Such an allelopathy-based technique for paddy weed control is the most easily transferable to the low-input management systems prevailing in most rice farming systems in Asia (Kong, 2008). Genetic modification of crop plants to improve their allelopathic properties and enhancement of their weed-suppressing ability has been suggested as a possibility. In agricultural production, breeding programme may give the possibility of utilizing rice allelopathy by inducing allelopathic traits into cultivated rice (Khanh et al, 2007). Certain rice cultivars have the potential to allelopathically suppress the seedling growth of barnyard grass. As a caution, before recommending an allelopathic cultivar for field trials, their effect on the N2-fixing potential of cyanobacteria should be studied in detail (Inderjit et al, 2001).
Diseases of rice (Oryza sativaL.) have threatened the stability of its production. Attempts to control these using synthetic biocides have met with limited success. Furthermore, the fate of these biocides is of great concern for agriculture, environment and human health. Several methods and techniques, including biological control, new resistant cultivars, natural biocides and allelochemicals, have been tried to improve rice disease management and control (Kong et al, 2010).
Allelochemicals are involved in practically every aspect of plant growth and can act as stimulating or suppressing agents. The smart utilization of allele-chemicals can be a major breakthrough in the agricultural sector (Khalid et al, 2002). Allelopathy can be used for biological control of pathogens to manage plant diseases. Several studies have successfully demonstrated the control of plant diseases using either allelopathic rotational crops or pure/crude allelochemicals (Rice, 1995).
Only few studies explored the allelopathic rice cultivars, which can deter or tolerate the insect pest pressure and pathogen attack. Screening of such resistant cultivars is highly desirable. The plant diseases can also be controlled by the antagonistic effects of antibiotics produced by other microorganisms (Rice, 1995).
Presence of phenolics and other allelochemicals in rice makes it a stronger competitor of insect pests and pathogens. Several lines of evidence indicate that momilactone A has an important role in rice defense system against pathogen attacks (Agrawal et al, 2002).
Among several diseases caused by bacterial, fungal, and viral pathogens that devastate rice yields all over the world, bacterial blight (Xanthomonas oryzaepv. oryzae), blast (Magnaporthe grisea), sheath blight (Rhizoctonia solani), sheath rot (Sarocladium oryzae), and tungro virus are the most important ones (Velusamy et al, 2006). X. oryzae pv. oryzae causes bacterial blight of rice and has high epidemic potential. It is destructive to high-yielding cultivars in both temperate and tropical regions especially in Asia. Certain plant-associated strains of fluorescent Pseudomonasspp. are known to produce the antibiotic 2, 4-diacetylphloroglucinol (DAPG) which inhibits the growth of X. oryzaepv. oryzaein laboratory assays and suppresses rice bacterial blight up to 59%-64% in net-house and field experiments (Velusamy et al, 2006). Two allelochemicals from rice, 5, 7, 40-trihydroxy- 30, 50-dimethoxy flavone (a flavone) and 3-isopropyl- 5-acetoxy cyclohexene-2-one-1 (acyclohexenone) are suggested to be a part of rice defense mechanism against diseases caused by two fungal pathogens (R. solani Kü hn and Pyricularia oryzaeCavara) (Singh et al, 2012).
In worldwide agricultural production system, the most important consideration is to improve the crop quality and yield which is mainly influenced by weeds (Khanh et al, 2007) and pests through environmentally safe agronomic approaches (Khanh et al, 2005). Application of allelopathy through genetic manipulation by using molecular genetics and biotechnology or conventional breeding in rice varieties can be considered as a successful tool for weed management, insect pests and disease pathogens (Bertin et al, 2008; Jabran and Farooq, 2013). Therefore, it is crucial to know which genes are responsible for allelopathic potential and also to understand the structure and genetic control of the allelopathic trait in order to incorporate the specific trait in rice breeding programs (Courtis and Olofsdotter, 1998). The genetic basis of allelopathy in rice by using F2 progeny derived from a cross between rice varieties PI312777 and Lemont revealed that allelopathy is quantitatively inherited (Dilday et al, 1998). Allelopathy in rice is polygenic and a quantitative trait which depends on several physiological and phenological characteristics (Kong et al, 2011). Jensen et al (2001) performed gene mapping and epistatic QTLs associated with allelopathic activity by using DNA markers and indicated that allelopathy in rice is a quantitative trait involving several loci and probably some levels of epistasis. Since several genes are involved in allelopathic activity, it may also affect the gene responsible for crop yield and thus it is very important to consider such aspect in breeding programmes. However, allelopathy in rice may be a trait that is not strongly associated with crop yield (Olofsdotter et al, 1995).
Due to evolution, natural and artificial selection traits including allelopathic traits have been incorporated in rice germplasm which is evidenced by the presence of certain degree of allelopathic activity even in non-allelopathic rice variety. Ebana et al (2001) reported seven QTLs related to rice allelopathy on chromosomes 1, 3, 5, 6, 7, 11 and 12, respectively, from F2 population of an indica-type line PI312777 and a japonica cultivar Lemont.
In order to understand the genetic control of allelopathy in rice, six genes namely DEG-1, DEG-4, DEG-5, DEG-7 (DEG-9) and DEG-8, with higher expression, and three genes, namely DEG-2, DEG-3 and DEG-6, with lower expression, were identified by using techniques like GeneFishing PCR and sequencing, when rice crop (Sathi) were grown with barnyard grass (Junaedi et al, 2008). These differential expressed genes responsible for allelopathic effect were further characterized (Table 3).
| Table 3 Genes and clones responsible for rice allelopathy. |
Lin et al (2005) utilized inter-simple sequence repeat as a molecular marker to assess the genetic diversity and cultivar differentiation of 57 rice accessions and 65 barely lines in terms of allelopathic potential. The study revealed that some accessions with higher allelopathic potential are grouped together, implying that the genes responsible for allelopathy might be isolocus, whereas some accessions of rice and barely with different allelopathic responses are also clustered into the same group, which perform low level of genetic polymorphism due to oriented selection for high yielding traits in breeding.
Rice allelopathy has been confirmed as an inducible genetic trait (Bi et al, 2007) that is associated with molecular regulation of the secondary metabolic pathways. Various secondary metabolites differentiated as allelochemicals are synthesized by a phenylpropanoid pathway. Some defense related proteins and the phenylalanine ammonia-lyase (PAL) enzyme, which is associated with phenylpropanoid metabolism and plant defense, have been found to be up-regulated in allelopathic rice (Fang et al, 2009). He et al (2005) used deferential proteomics and bioinformatics to study the molecular mechanism of rice allelopathy grown with barnyard grass. They found that the enhanced inhibitory effect of allelopathic rice on target weeds was due to the up-regulated expression of gene which encodes PAL.
Song et al (2008) classified 24 genes into 5 groups i.e. primary metabolism, phenolic allelochemical synthesis, plant growth/cell cycle regulation, stress response/signal transduction, and protein synthesis/ degradation based on their functions (Table 3). Further, up-regulation of the putative genes that encode for PAL, o-methyltransferase, triosephosphate isomerase and cytochrome P450, which are involved in de novo synthesis of phenolic allelochemicals and detoxification of toxic substances in PI312777 (allelopathic rice), was detected by subtractive hybridization suppression at a low nitrogen supply. Similarly, RNAi was used to inhibit PAL expression in allelopathic rice accession PI312777 by Fang et al (2013) in order to confirm the role of PAL gene in the regulation of rice allelopathy with respect to synthesis, release and metabolism of phenolics. It was observed that the down-regulation of PAL gene expression decreases the gene expression of phenolic metabolism-related enzymes and lowers the level of phenolics that in turn reduces the allelopathic potential of rice on barnyard grass. Reduction of phenolic exudates in transgenic rice also influences the quantity of rice rhizospheric microbes with eight phyla lesser in transgenic plants compared to wild type (Fang et al, 2013). Also, Lin et al (2011) and Cipollini et al (2012) found that phenolic acids are useful carbon resources that help establish the soil microbial community. The application of phenolics may improve microbial population in paddy soil and stimulate the activities of soil microorganisms which play a vital role in enhancing the allelopathic effect. The accumulation of PAL mRNA induced by exogenous salicylic acid will stimulate the synthesis of new PAL protein, increasing PAL enzyme activity against pest and weed infection in crops. Therefore, salicylic acid can be used as an activator to induce allelopathic defense in rice, especially in allelopathic rice (Bi et al, 2007; Fang et al, 2009). It has been generally assumed that the appearance of phenylpropanoid metabolites during a plant’ s response to weed and pest infection is a result of the transcriptional activation of the various biosynthetic pathway genes (Dixon, 2002). Lin et al (2004) performed proteome analysis i.e. MALDI-TOF/MS to investigate the different expression of proteins in allelopathic rice exposed to barnyard grass stress. This study provided four induced proteins, 3-hydroxy- 3-methylglutaryl-coA reductase 3 (HMGR3), PAL, thioredoxin-m and peroxidase precursor, which are related to the pathways of isoterpenoid and phenylpropanoid biosynthesis in the plant defence response. Therefore, it could be considered that proteomic approach can contribute to the identification of positional, functional and expressional genes. Comparison of 2-DE protein pattern obtained for key tissue of stressed and control plants will identify a set of stress-responsive proteins encoded by expressional candidate genes. Sequencing of these stressed- responsive proteins will then reveal that some of them have functions clearly consistent with the stress tolerant traits. The encoding genes will thus be both the expressional and functional candidate genes.
Beside such intensive information about allelopathic genetics, very less breeding efforts (especially with respect to disease management) have been made to exploit allelopathy as commercially acceptable biocontrol method in modern agricultural practice. In order to get best results of hybrid varieties, it is very important to consider the allelopathic effect of donor against wide spectrum of harmful biological system (like weeds, pests and fungi). However, while executing such an idea, the main concern should be the impacts of such improved crops on crop yield, other beneficial organisms, soil, environment, the residue, the next year crop, etc.
Since its inception in 1937, the term allelopathy is mainly viewed in terms of negative communications, but latter it was proved that, if correctly managed, this phenomenon may be exploited for enhancing the crop productivity. The number of reports indicating the improvement in crop production due to allelopathic interactions is increasing. This manipulation can be achieved by weed management, disease management, pest management, nitrogen management, etc. For sustainable agriculture of rice, allelopathy has achieved great success in weed management. Utilization of water extracts of allelopathic crop combined with reduced doses of herbicides can be a promising strategy for sustainable weed management and environment health.
The allelopathic potential of crop can be exploited directly by using allelopathic interactions or indirectly by utilizing allelochemicals as biopesticide. About 16 000 rice accessions from 99 countries have been screened for their allelopathic potential against different weeds. Use of rice residues for weed control in paddy fields is traditionally used by many farmers across the globe, however, it requires large amount of biomass.
Although there are several promising rice allelochemicals reported to inhibit weed growth and some pathogenic organisms, their direct use as pesticides is not successful due to several reasons, viz. their stability under natural environment, selectivity and limited activity, effect on non-target organisms, etc. Also, it is very difficult and expensive to develop new novel compounds to be used as pesticides. Even the isolation of allelochemicals from plants in required amount is a tedious process. Therefore, searching a compound with simple chemical structure (which might reduce its synthesis cost) and strong activity is required in future as it has achieved limited success for commercial production.
A regulation of the biosynthesis and the release rate to enhance the flow of allelochemicals (hopefully non-toxic to humans) or to prolong the period of release of the allelochemicals has been suggested (Wu et al, 1999). Use of biotechnological transfer of allelopathic traits between cultivars of the same species or between species has also been proposed (Chou, 1999), since it is easy and efficient to screen for the presence of allelopathic characteristics by using several molecular markers.
An apparently untouched aspect of allelopathy is its effect on the nutritional value of food and fodder crops. However, a change in the combinations or levels of the secondary metabolites in crops should not affect their quality as food products. Finally, the structure of allelochemicals can be used as an analogue for the synthesis of new pesticides. These biopesticides will perhaps be far less harmful for the environment as compared to synthetic agrochemicals.
However, it is very essential to consider some key aspects before applying allelopathy in natural conditions, like: the duration of allelopathic activity (stability and persistence of allelochemicals), synergistic interactions by studying threshold concentration of each compound and the behavior of genes in cross pollination conditions.
The authors have declared that no competing interests exist.
| 1 |
|
| 2 |
|
| 3 |
|
| 4 |
|
| 5 |
|
| 6 |
|
| 7 |
|
| 8 |
|
| 9 |
|
| 10 |
|
| 11 |
|
| 12 |
|
| 13 |
|
| 14 |
|
| 15 |
|
| 16 |
|
| 17 |
|
| 18 |
|
| 19 |
|
| 20 |
|
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|
| 25 |
|
| 26 |
|
| 27 |
|
| 28 |
|
| 29 |
|
| 30 |
|
| 31 |
|
| 32 |
|
| 33 |
|
| 34 |
|
| 35 |
|
| 36 |
|
| 37 |
|
| 38 |
|
| 39 |
|
| 40 |
|
| 41 |
|
| 42 |
|
| 43 |
|
| 44 |
|
| 45 |
|
| 46 |
|
| 47 |
|
| 48 |
|
| 49 |
|
| 50 |
|
| 51 |
|
| 52 |
|
| 53 |
|
| 54 |
|
| 55 |
|
| 56 |
|
| 57 |
|
| 58 |
|
| 59 |
|
| 60 |
|
| 61 |
|
| 62 |
|
| 63 |
|
| 64 |
|
| 65 |
|
| 66 |
|
| 67 |
|
| 68 |
|
| 69 |
|
| 70 |
|
| 71 |
|
| 72 |
|
| 73 |
|
| 74 |
|
| 75 |
|
| 76 |
|
| 77 |
|
| 78 |
|
| 79 |
|
| 80 |
|
| 81 |
|
| 82 |
|
| 83 |
|
| 84 |
|
| 85 |
|
| 86 |
|
| 87 |
|
| 88 |
|
| 89 |
|
| 90 |
|
| 91 |
|
| 92 |
|
| 93 |
|
| 94 |
|
| 95 |
|
| 96 |
|
| 97 |
|
| 98 |
|
| 99 |
|
| 100 |
|
| 101 |
|
| 102 |
|
| 103 |
|
| 104 |
|
| 105 |
|
| 106 |
|
| 107 |
|
| 108 |
|
| 109 |
|
| 110 |
|
| 111 |
|
| 112 |
|
| 113 |
|
| 114 |
|
| 115 |
|
| 116 |
|
| 117 |
|
| 118 |
|
| 119 |
|
| 120 |
|
| 121 |
|
| 122 |
|
| 123 |
|
| 124 |
|
| 125 |
|
| 126 |
|
| 127 |
|
| 128 |
|
| 129 |
|
| 130 |
|
| 131 |
|
| 132 |
|
| 133 |
|
| 134 |
|
| 135 |
|
| 136 |
|
| 137 |
|
| 138 |
|
| 139 |
|
| 140 |
|
| 141 |
|
| 142 |
|
| 143 |
|
| 144 |
|
| 145 |
|
| 146 |
|
| 147 |
|
| 148 |
|
| 149 |
|
| 150 |
|
| 151 |
|
| 152 |
|
| 153 |
|
| 154 |
|
| 155 |
|
| 156 |
|
| 157 |
|
| 158 |
|
| 159 |
|
| 160 |
|
| 161 |
|
| 162 |
|
| 163 |
|
| 164 |
|
| 165 |
|
| 166 |
|
| 167 |
|
| 168 |
|
| 169 |
|
| 170 |
|
| 171 |
|
| 172 |
|
| 173 |
|
| 174 |
|