The waxy gene ( Wx) in rice, which encodes the granule bound starch synthase enzyme, is responsible for amylose synthesis. Glutinous (sticky) rice has little or no amylose that can be used in various applications, such as brewing. In this study, knockout of the Wx gene with CRISPR/Cas9 technology was conducted in two elite japonica rice lines, Huaidao 5 (HD5) and Suken 118 (SK118), aiming to develop elite sticky rice varieties. We achieved six homozygous T0 plants with more than 200 bp deletion in the Wx gene, as well as 36 wx-HD5 and 18 wx-SK118 homozygous transgene-free plants in the T1 generation. The seeds of all the mutants were white and opaque, similar to those of sticky rice, and contained only 2.6%-3.2% amylose. Results of scanning electron microscopy showed that the quality of rice did not change. In conclusion, we successfully developed two elite sticky rice varieties.
Rice is one of the most important crops globally. High yield, good quality and resistant traits are important targets in its breeding programs (Kiswara et al, 2014), and genetic diversity in rice germplasm is crucial for these purposes. Various strategies, such as exploring wild rice germplasm and natural or artificial mutation, have been employed by breeders to broaden the genetic diversity of rice (Shen et al, 2017). However, these methods are time consuming, and in the case of gene mutations, randomly. The clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 (CRISPR/Cas9) system is a novel technology, widely used in various fields because of its cost effective, simple, flexible and highly efficient (Wang et al, 2017a). In rice, numerous studies have been performed with the CRISPR/Cas9 system (Wang et al, 2016). Ma et al (2015) reported that editing efficiency of CRISPR/Cas9 is higher in rice than in Arabidopsis. Mikami et al (2015) found that the OsU6 promoter performs better than the OsU3 in mutation frequency. A 1-bp mismatch of an sgRNA is sufficient to avoid off-target in closely related genes in rice (Baysal et al, 2016). There is a greater probability of large gene deletions in japonica rice than in indica rice (Wang et al, 2017b). With the rapid development of the CRISPR-Cas system, many researchers have begun to use this technology to improve agronomic traits in rice. For example, rice blast resistance lines are produced by CRISPR/Cas9-targeted mutagenesis of OsERF922 (Wang et al, 2016). Hybrid-compatible lines are developed by knocking out the Sa genes (Xie et al, 2017). Low-Cs+/Cd rice strains are also created by knocking out the corresponding metal transporter genes (Nieves-cordones et al, 2017; Tang et al, 2017).
Amylose is a linear polymer composed of glucose units, linked by α -D-(1-4) glycosidic linkages. In rice, amylose content (AC) is a complex trait, influenced by multiple genes, mainly the waxy (Wx) gene, and the environment (Chen et al, 2008). The Wx gene encodes the granule-bound starch synthase I (GBSSI), which catalyzes the synthesis of amylose. The expression of Wx is modulated by many factors, which can lead to a change in AC (Wang et al, 2015). Wu et al (2015) reported that dozens of dull genes regulate the splicing efficiency of Wx. A tetratricopeptide domain-containing protein (flo2) regulates the expression of Wx (Wu et al, 2015). rsr1, OsBP-5, OsEBP-89, OsbZIP58 and OsMADS7, as transcription factors, modify Wx expression (Wamnugu et al, 2017). Many QTLs, such as qAC2, have been identified as genes controlling AC (Fasahat et al, 2014). Glutelin, gibberellin receptor, ADP- glucose pyrophosphorylase and pullulanase also play minor roles in affecting AC (Wamnugu et al, 2017). Here, we used CRISPR/Cas9 technology to edit the Wx gene and generate two elite glutinous rice varieties that could meet the demands of consumers and enrich the rice genetic resources.
Rice (Oryza sativa) inbred lines Huaidao 5 (HD5) and Suken 118 (SK118) were used for Agrobacterium-mediated co-cultivation transformation experiments, as described previously (Zheng et al, 2016). During growing seasons, all the materials were cultivated in the greenhouse of the Jiangsu Academy of Agricultural Sciences in Nanjing, Jiangsu Province, China.
| Table 1 Primers used in this study |
The Wx gene (LOC_Os06g04200) was the target of Cas9 endonuclease. According to the principles outlined in Ma et al (2015), two sgRNAs (T1 and T2) were designed on the first extron of the Wx gene from Nipponbare. The sgRNAs were assembled into one vector pYLCRISPR/Cas9-MH using BsaI (New England Biolabs, Country) and T4 ligase (New England Biolabs, Country). T1 and T2 were driven by OsU3 and OsU6a promoters, respectively. The vector was transformed into the Escherichia coli strain DH5α , and the insertions were verified by sequencing. The primers sequences of the sgRNAs are listed in Table 1. A vector map was generated using vector NTI (Lu and Moriyama, 2004).
The genomic DNA of transgenic plants was isolated from rice leaves using the CTAB method (Murray and Thompson, 1980). The PCRs were conducted using a Golden Star T6 Super PCR Mix (TsingKe Biotech Co., Ltd, China) to amplify the sequences of T1 and T2. The reagents and reaction conditions of the PCRs were selected and programmed according to the manufacturer’ s instructions. The obtained DNA fragments were sequenced, and then a similarity search using BLAST was performed with the wild-type (WT) Wx sequence.
To identify the transgene-free lines, T1 seedlings were examined using PCR with specific primers (Table 1) for the sequences of Cas9 and hygromycin-resistance gene (HPTII).
The rice seeds were harvested at maturity and dried in an oven at 38 º C for two days after manual dehulling. The morphology of the seed endosperms was observed by visual inspection. Potassium iodide (I2-KI, 2%) staining was used to identify the starch types in the transgenic seeds T1 generation (Kawagoe et al, 2005). Dried grains were ground into fine flour to pass through a 60 mesh sieve. Then, the AC of the powder was measured using a colorimetric method (Juliano et al, 1981). Scanning electron microscopy (SEM) was also performed to investigate the morphology of the rice starch. The polished rice seeds were cut with the blade of a scalpel, mounted on copper stubs, and coated with gold. All detailed procedures were performed according to Kasem et al (2011). Images were recorded on a Hitachi-S-3000N scanning electron microscope (Hitachi, Co., Ltd. Japan).
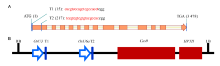
To delete the Wx gene in japonica rice with the CRISPR/Cas9 technology, two different 20-bp sgRNAs (T1 and T2) at the first exon of Wx were selected using the web-based tool CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR), according to the criteria reported by Ma et al (2015). Two target sites were located at 15 and 217 bp downstream of the translation initiation codon (ATG) and were named T1 and T2, respectively (Fig. 1-A). The length between T1 and T2 was 202 bp, according to the coding sequence of Wx. Two sgRNAs were sequentially assembled into pYLCRISPR/Cas9-MH as described by Wang et al (2017c). The T1 and T2 in the plasmid were driven by the OsU3 and OsU6a promoters, respectively. Both Cas9 and HPTII, elements of the plasmid, were transcribed from the CaMV 35S promoter (Fig. 1-B).
The CRISPR/Cas9-Wx construct was transformed into two elite rice inbred lines HD5 and SK118. We obtained 36 and 32 independent T0 transgenic plants of HD5 and SK118, respectively. The PCR tests of the HPTII gene confirmed that all T0 plants were positive transgenic lines (Fig. 2-A). To analyze the mutation genotypes of the T0 plants, three mutants of both wx-HD5 and wx-SK118 were randomly selected for PCR amplification (Fig. 2-B). The PCR products were sequenced. The genotype differences between the T0 mutants and the WT lines are shown in Fig. 2-C. The results showed that the mutagenesis efficiency of the target sites was very high, the deletion fragments were larger than 200 bp in all individuals, the nucleotides between two protospacer adjacent motif (PAM) sites of the targets were all deleted by CRISPR/Cas9, and all T0 plants were homozygous mutations.
Because inherited Cas9 could also induce new mutations and lead to unpredicted segregation, distortion and chimerism in later generations, it is important to eliminate Cas9 by segregation in the T1 generation and obtain stable mutants (Ishizaki, 2016; Yin et al, 2017). Three wx-SK118 (wx-SK118-1, wx-SK118-2 and wx-SK118-3) and three wx-HD5 (wx-HD5-1, wx-HD5-2 and wx-HD5-3) T0 mutants were self-pollinated to generate T1 plants. Transgene-free lines were analyzed by PCR using specific primers for the Cas9 and HPTII sequences in the T1 plants (Table 1). The results were determined using negative PCRs of both Cas9 and HPTII. The 235 wx-HD5 and 118 wx-SK118 T1 plants (including selections from all the six lines) were subjected to PCR tests, respectively, and 36 wx-HD5 and 18 wx-SK118 transgene-free plants were isolated, respectively (Table 2).
| Table 2 Segregation of transgene-free rice in T1 generation. |
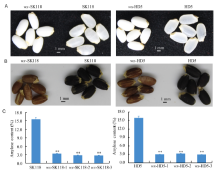
The phenotypes of the mutant grains were compared with those of the WT. As shown in Fig. 3-A, the grains of the mutants were all white and opaque, which showed similar characteristics to those of typical glutinous rice varieties. The starch properties of the mutants were identified by a screen with I2-KI staining, as the seeds with more amylose have higher iodine binding capacity. Non-glutinous rice varieties have granules that are stained with a dark blue color. Seeds with more amylopectin, such as sticky rice, have a lower iodine binding capacity, and therefore, granules are stained with a reddish brown color (Kawagoe et al, 2005). As shown in Fig. 3-B, all the seeds of the mutants were reddish brown, while the WT grains were dark blue. The results revealed that the amylose content in the mutants was dramatically decreased. Further analysis showed that the AC of the mutants was 2.6%-3.2%, while the AC of the WT grains was 15.4%-15.7% (Fig. 3-C). As expected, the dramatic decrease of AC in the mutants was closely associated with the grain appearance and iodine staining results. The wx-HD5 and wx-SK118 rice strains were turned into glutinous lines.
The cross sections of the grains were observed with SEM (Fig. 4). Results in Fig. 4-A revealed that the grain surfaces of the mutants were smoother and flatter than in the wild-types. Radiation from the rectangular columns, as revealed by cross-sections of grains, was reduced in the mutants, especially in wx-HD5. The fractured surfaces of grains from both the mutants and WTs were all densely packed with starch granules, and all the starch granules displayed large and irregular polygonal shapes with sharp edges (Fig. 4-B).
The first application of CRISPR/Cas9-based genome editing in rice was presented in 2013 (Cho et al, 2013). Since then, the technology has been widely used to improve agronomic traits and create novel germplasm in rice. Li et al (2016) and Zhou et al (2016) developed photoperiod-controlled and thermo-sensitive male sterile lines by disrupting the CSA and TMS5 genes, respectively. Mutants of the GW2, GW5 and TGW6 genes, which function as negative regulators of grain weight, were generated through CRISPR/Cas9-mediated targeted mutagenesis (Xu et al, 2016). Li et al (2017) designed sgRNAs to target Hd2, Hd4 and Hd5, and produced early-maturing rice germplasm, which can improve the ecological adaptability of those rice varieties. Sun et al (2017) created high amylose rice plants through CRISPR/Cas9-mediated target gene editing of SBEIIb and SBEI. The CRISPR/Cas9 technology has a great potential to facilitate plant breeding. Given this, we developed two elite glutinous rice varieties using the CRISPR/Cas9 technology by editing the Wx gene.
We designed two sgRNAs (T1 and T2), targeting the first exon of the Wx gene. The GC contents of T1 and T2 were 70% and 65% respectively, and the secondary structures of sgRNAs had less than 6 bp complete matching. Using these sgRNAs, we observed extremely high editing efficiency in the Wx gene. The design parameters of the sgRNA are in line with the reported critia (Ma et al, 2015), which might be a reason for the high sgRNA editing efficiency in this study. It was very interesting that the genotypes of all mutants generated from six lines were homozygous with more than 200 bp deletions and that the fragment between T1 and T2 was missing. It has been reported that CRISPR/Cas9 induces modifications that are small, presumably because of the blunt-ended double-strand breaks (Zheng et al, 2016). In this study, the CRISPR/Cas9 systerm seemed to preferentially induced full-length deletions, rather than small modifications. When the number of base pairs between the two target sites in Wx was 202 bp, and so that we found that the genetypes of all the six selected T0 transgenic plants were about 200 bp-fragment deletion. A similar phenomenon was described by (Wang et al, 2017a), but the distance between the sgRNAs required to induce large deletions should be further explored.
Wx mutation with different amylose content has been reported (Toru and Naoko, 2017). Rice varieties can be classified as waxy (0-2%), very low (3%-9%), low (10%-19%), intermediate (20%-25%) and high (> 25%) according to their AC (Cai et al, 2015). The Wx gene is the principal gene controlling the AC and can explain almost 90% of the AC variation in some cultivars (Wamnugu et al, 2017). Various Wx mutations (Wxa, Wxb, Wxmq, Wxmp, Wxin and wx) generate a wide range of variation in AC (Yang et al, 2016; Wang et al, 2018). Studies have reported that different types of Wx mutations, such as a G/T mutation at the 5′ splice site of intron 1, a C deletion at intron 5, a G/A SNP in the Wx promoter region, or a T/G SNP at the intron 1-exon 1 junction, would result in amylose content decreased (Wang et al, 2015; Shahid et al, 2016). Ma et al (2015) disrupted the Wx gene by CRISPR/Cas9 system in a japonica rice Taichung 65, and the AC in the mutants decreased to 2.6%, which is similar to a natural glutinous rice. Zhang et al (2018) used the CRISPR/Cas9 to target the first exon of Wx in two japonica rice varieties, Xiushui 134 and Wuyunjing 7, and the results showed that the AC of mutants is reduced without affecting the other desirable agronomic traits. Our results generally agree with those reported in other studies.
In conclusion, we precisely edited the Wx gene using CRISPR/Cas9 technology and generated high-quality glutinous rice. This work would provide new materials for traditional breeding and broaden the genetic diversity of glutinous rice.
This work was supported by the National Key Research and Development Program (Grant No. 2017YFD0100403), the Jiangsu Province Key Research and Development Program (Modern Agriculture) Project (Grant No. BE2017345-2), the Exploratory Project of the Jiangsu Academy of Agricultural Sciences [Grant No. ZX(17)2014], and the Jiangsu Province Natural Science Foundation (Grant No. BK20171326).
The authors have declared that no competing interests exist.
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|