SUMMARY Precise base editing is highly desired in plant functional genomic research and crop molecular breeding. In this study, we constructed a rice-codon optimized adenine base editor (ABE)-nCas9 tool that induced targeted A∙T to G∙C point mutation of a key single nucleotide polymorphism site in an important agricultural gene. Combined with the modified single-guide RNA variant, our plant ABE tool can efficiently achieve adenine base editing in the rice genome.
The CRISPR/Cas9 system has been successfully employed in targeted genome mutagenesis in rice and various plant species, providing a promising strategy to accelerate crop improvement. The system is generally used to create gene knockouts or loss-of-function mutations by inducing the random deletions or insertions through the error-prone non-homologous end joining (NHEJ) mechanism. However, a number of agricultural traits are conferred by single nucleotide polymorphisms (SNPs). Despite extensive efforts in developing reliable point mutations or base replacement methods through the homology-directed repair (HDR) mechanism (Miki et al, 2018), it is haunted by a troublesome fact that the low amounts of donor DNA templates limit the efficiency of precise gene editing in plant cells. Base editing is a novel approach that induces single base conversion without requiring HDR or DNA double-strand breaks (DSBs) (Komor et al, 2016). The most well-known base editor tool, base editor 3 (BE3), is consisted of an nCas9 (Cas9-D10A nickase), the rat APOBEC1 cytidine deaminase, and the uracil DNA glycosylase inhibitor (UGI) (Komor et al, 2016). Cooperating with single-guide RNA (sgRNA), the nCas9 fusion protein is directed to the target genome site, and efficiently induces C to T or G to A substitutions (Komor et al, 2016). The BE3 system is highly capable of precise crop breeding by mediating C∙ G to T∙ A conversion at key functional SNP loci associated with various important agricultural traits, such as herbicide resistance, plant type and nitrogen use efficiency (Lu and Zhu, 2017; Zong et al, 2017).
A∙ T to G∙ C mutations frequently occur as functional SNPs. Recently, a novel base editing tool, the adenine base editor (ABE), was developed by extensive protein engineering based on the bacterial tRNA adenosine deaminase (TadA) (Gaudelli et al, 2017). Natural TadA converts A to inosine (I) in tRNA but not DNA. After several rounds of protein evolution, several Escherichia coli TadA (ecTadA) mutants (TadA* ) with DNA- modifying capabilities were created (Gaudelli et al, 2017). To assemble the ABE, a heterodimer consisting of TadA* and wild-type TadA was formed into a modified Cas9 enzyme. In mammalian cells, the late-stage ABE tool, e.g., ABE7.10, can be programmed to induce A∙ T to G∙ C conversion at target loci without undesired InDels or off-target effects (Gaudelli et al, 2017). In this study, we reported the efficient A∙ T to G∙ C conversion in the rice genome using the ABE7.10 system.
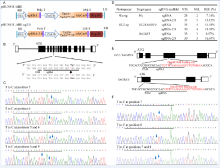
We first synthesized the rice codon-optimized ecTadA- XTEN-TadA* 7.10, which was then attached to the N-terminal SpCas9 (D10A) nickase (Supplemental Fig. 1). The sequence of the recombinant protein was cloned to our previously modified pHUN411 binary vector under the control of a maize ubiquitin promoter. This vector, designated as pHUN411-ABE (Fig. 1-A), still contains a rice U3 promoter cassette for protospacer sequence integration and sgRNA expression (Xing et al, 2014).
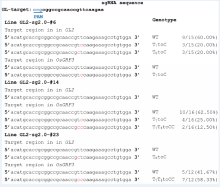
The vector was tested in the rice amylose synthesis gene Wx. Wx-mq is a minor mutant allele that results in low amylose content in rice endosperm (Sato et al, 2002). The Wx-mqallele contains a T to C substitution at position 595, resulting in the replacement of a tyrosine by a histidine at residue 191. To mimic this mutation, a sgRNA (Wx-sg) was designed (Fig. 1-B) and constructed into the pHUN411-ABE vector. The construct was transformed into rice plants via Agrobacteria, and 28 independent transgenic events were regenerated. The target mutants were screened by high-resolution melting analysis and then confirmed by PCR sequencing. Two targeted mutants were identified (Supplemental Table 2). One line (Wx-6, the 2nd chromatogram) harbored a T to C substitution at position 6 (the first nucleotide of the 20-bp sgRNA distal from PAM is position 1) (Fig. 1-C), whereas the other line (Wx-15, the 3rd chromatogram) had two T to C mutations at positions 5 and 6 (Fig. 1-C). The genotype of the Wx-15 line was further assessed by clone sequencing. We found that approximately 16.67% clones had the desired T to C conversion at position 5, and approximately 27.78% clones harbored the substitution at position 6 (Supplemental Fig. 2). To increase editing efficiency, an extended version of the sgRNA was used to replace the conventional sequence in pHUN411-ABE (Supplemental Fig. 3) (Dang et al, 2015), generating the pHUN411-ABE-sg2.0 vector. The Wx-sg was fused into the ABE-sg2.0 binary vector, and base changes were observed in 5 out of the 33 transgenic lines (Fig. 1-D). Among these lines, Wx-sg2.0-7 and Wx-sg2.0-22 were edited at positions 5 and 6, respectively. Three lines (Wx-sg2.0-2, Wx-sg2.0-15 and Wx-sg2.0-21) were potentially chimeric (Supplemental Table 2). After detailed genotyping, Wx-sg2.0-21 showed that the T at positions 5 and 6 was simultaneously mutated to C in a single strand (Supplemental Fig. 2). Furthermore, a T to C substitution at position 9 was detected in Wx-sg2.0-15 (Fig. 1-C and Supplemental Fig. 2), although the editing window of ABE was believed to be limited to 4 bases, ranging from positions 4 to 7 (Gaudelli et al, 2017).
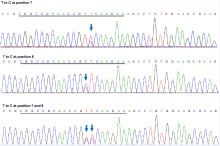
To further test the editing feature of the ABE vectors in rice, we targeted the GL2/OsGRF4 and OsGRF3 genes, which is responsible for grain size and yield. It has been shown that the 2-bp substitution mutation located in the miR396 recognition sequence of GL2 disrupts miR396-directed GL2 transcript cleavage, resulting in heavier grain weight and higher yield (Che et al, 2015). Fig. 1-E shows that the miR396 binding site in GL2 was targeted by sgRNA GL2-sg. This protospacer also exhibits 100% sequence identity to the miR396-binding region of OsGRF3(Fig. 1-E) (Liu et al, 2014). We found that 4 out of the 35 transgenic lines of the pHUN411-ABE-GL2-sg harbored the targeted base mutations (Fig. 1-D and Supplemental Table 3). Among these, three lines had the substitution in the target region of both GL2 and OsGRF3, whereas one harbored the mutation in GL2 but not OsGRF3(Supplemental Table 3). At the target region in GL2, three lines carried the T to C substitution at position 7, and the T to C substitution at position 8 was only observed in one transgenic event (Fig. 1-F and Supplemental Table 3). In addition, we observed that the mutation only occurred at position 7 in the target of OsGRF3 (Supplemental Figs. 3 and 4, and Supplemental Table 3), suggesting that the targeting feature of the ABE tool has position preference for the editing window. The protospacer was also fused to the extended sgRNA variant, and 4 out of 30 transgenic lines harbored the targeted base mutation in the GL2 sequence (Fig. 1-D and Supplemental Table 3). Furthermore, editing in the target region of OsGRF3 was also detected in five independent lines (achieving 16.67% efficiency), which included three lines that were targeted at both sites (Fig. 1-D and Supplemental Table 3). In most of the mutated plants, the conversion of nucleotide T at positions 7 and/or 8 into C was heterozygous or chimeric (Supplemental Fig. 5 and Supplemental Table 3). Off-target effects of the plant ABE tool were also detected. The most frequent off-target region in GL2-sg was located in the miR396-binding site of OsGRF5, with a single base mismatch (Supplemental Fig. 6) (Liu et al, 2014). Assessment of all transgenic plants using both vectors showed no off-target mutations (Supplemental Table 4).
In this study, we reported the efficient A∙ T to G∙ C base editing in rice. The A∙ T to G∙ C mutation can result desired amino acid substitution or potential interference of miRNA binding in the target regions (Supplemental Fig. 7). We noticed the mutation frequency induced by the pHUN411-ABE vector was < 10% at the Wx and GL2 targets. A possible explanation is that a high resolution method (HRM) analysis was used to pre-screen the target mutations, which may ignore some lines with low-frequency chimeric mutations. In plants, the mutagenesis efficiency of the Cas9 variant SpCas9-VQR could be significantly increased by the adapted sgRNA with extended Cas9-binding hairpin structure and mutations on the potential terminator of the pol III promoter (Hu et al, 2018). Using this version of the sgRNA variant, the base editing efficiency of the Cas9-ABE tool was substantially increased, suggesting that the modification of the sgRNA structure improves the efficiency of the ABE tool. Three groups also most recently and independently described a similar A∙ T to G∙ C base editing by ABE in plants (Hua et al, 2018; Li et al, 2018; Yan et al, 2018). The rBE14 system exhibits 16.67% to 62.26% editing efficiency at three different genomic loci in rice (Yan et al, 2018). The pRABEsp-OsU6 system could achieve as high as 26% editing efficiency in the T0 rice transgenic population (Hua et al, 2018), and the PABE-7 system, developed by a series optimization on position of TadA, nuclear localization sequences locations, and also sgRNA expression, strategies exhibits A to G substitution efficiency of 15.8% to 59.1% in the stable transformed rice lines (Li et al, 2018). The nucleotide sequence context might be a major contributing factor to the efficiency variations of different SpCas9-based plant ABE tools. It will be interesting to test them, including our ABE system, in the same target loci under the same transformation procedure. Furthermore, a recent study in mammalian cells indicated that base editing efficiency could be affected by multiple factors during engineering, including the different commercial source of codon-optimization (Koblan et al, 2018). Taken together, the plant ABE systems greatly expand the application of CRISPR-Cas9 tools as well as provide a reliable and efficient method for fundamental plant research and precise crop breeding.
We appreciate Prof. Chen Qijun for providing the pHUN411 vector. This study was supported by the National Natural Science Foundation of China (Grant No. 31501239) and the Genetically Modified Breeding Major Projects (Grant No. 2016ZX08010-002-008).
The following materials are available in the online version of this article at http://www.sciencedirect.com/science/journal/16726308; http://www.ricescience.org.
Supplemental File 1. Materials and methods used in this study.
Supplemental Table 1. The primers used in this study.
| Supplemental Table 1 The primers used in this study. |
Supplemental Table 2. Identification of targeted wxmutants induced by adenine base editor tools in transgenic rice.
| Supplemental Table 2 Identification of targeted wxmutants induced by ABE tools in transgenic rice |
Supplemental Table 3. Identification of targeted gl2and/orosgrf3mutants induced by adenine base editor tools in transgenic rice.
| Supplemental Table 3 Identification of targeted gl2and/orosgrf3mutants induced by ABE tools in transgenic rice. |
Supplemental Table 4. Detection of the putative off-target effects of the rice adenine base editor tools on the high-similarity site of the GL2-sg protospacer.
| Supplemental Table 4 Detection of the putative off-target effects of the rice ABE tools on the high-similarity site of the GL2-sg protospacer. |
Supplemental Fig. 1. Sequence of rice codon-optimized ABE7.10.
 | Fig. 1. Sequence of rice codon-optimized ABE7.10. The sequence of wild type ecTadA, linker, TadA* 7.10, nCas9(D10A) and NLS was labeled by blue, orange, red, black and cyan respectively. |
Supplemental Fig. 2. Genotyping of some targetedwx mutants.
Supplemental Fig. 3. The sequence of the extended sgRNA scaffold.
 | Fig. 3 The sequence of the extended sgRNA scaffold. The structures of the sgRNA-scaffold-2.0 sequence were generated with RNAfold and visualized using VARNA (Darty et al., 2009). The adapted and extended RNA is shown in red. |
Supplemental Fig. 4. Sanger sequencing chromatograms of the OsGRF3 target region of representative ABE-Gl2-sg edited lines.
Supplemental Fig. 5. Genotyping of some transgenic plants targeted by ABE-GL2-sg.
Supplemental Fig. 6. Alignment of the miR396 binding region in the rice growth-regulating factor family.
Supplemental Fig. 7. The mutation of protein sequence resulting from the adenine base editor-induced editing.
The authors have declared that no competing interests exist.
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|