As a main feature of plant autotrophy, assimilation of inorganic nitrogen (N) is not only of fundamental interest to the crop, but also a crucial factor in crop productivity. N is the main plant mineral nutrient needed for chlorophyll production and other plant cell components (proteins, nucleic acids and amino acids). I highlighted the novel aspects of N responsive sensors, transporters and signaling molecules recently identified in the monocot rice plant, and discussed their potential roles in N sensing and transporting. Furthermore, over the last couple of years, N sensing has been shown to be affected by different external factors, which act as local signals to trigger systemic signaling coordinated by long-distance transport or mobile signals in plant body. Understanding of this complex regulatory network provides a foundation mechanism for the development of novel strategies to increase the acquisition and transportation efficiency of nitrogen under varying N conditions for rice production.
As a mineral nutrition, most terrestrial organisms (fungi, bacteria and plants) rely on the uptake of inorganic ions from the soil or terrestrial environment. In the natural ecosystems, the availability of these ions dramatically fluctuates in both time and space, which makes nutrient-limiting conditions. To face this natural constraint, all organisms develop adaptive responses triggered by sensing systems that perceive external nutrient availability (Holsbeeks et al, 2004; Schachtman and Shin, 2007; Gojon et al, 2009). Plants are non-motile and therefore they have evolved a highly sophisticated and complex sensing and signaling mechanisms to respond to the dynamic changes of their surrounding environments. After carbon, hydrogen and oxygen, nitrogen (N) is one of the essential elements in plants due to its key role in chlorophyll production and is fundamental for the photosynthesis process. In addition, N is a part of various enzymatic proteins that catalyze and regulate plant-growth processes (Sinfield et al, 2010). Furthermore, N contributes to the production of chemical components that protect against parasites and plant diseases (Hoffland et al, 2000). Finally, crop yield and biomass are highly affected by N fertilization (Tremblay et al, 2011). N as a mineral nutrient is absorbed by plants mainly from soil, and it can be in the forms of ammonium (NH4+) and nitrate (NO3-) (Taiz and Zeiger, 2010). However, the supply N in soil is often limited (Vigneau et al, 2011), which forces the farmers to increase the application of N fertilizers in order to achieve high crop yield. Increasing demand for rice as a staple food, the requirement for environmentally friendly agriculture and future risks arising from climate change are all associated with the urgent need to improve N use efficiency (Zhang et al, 2015). However, farmers may provoke over fertilization especially for N, which thwarts optimum productivity of rice plant (Rubio-Covarrubias et al, 2009), as rice plants are not able to absorb the excess N-fertilizer. This entails unnecessary expenditure on the part of farmers. The nutrition of nitrogen in rice plants is milestone allowing the associated, interdisciplinary scientific community to exchange ideas on recent findings concerning the mechanisms of inorganic N assimilation by plants.
To date, nutrient sensors for different minerals are mostly uncharacterized in rice plants (Schachtman and Shin, 2007), but recent findings in Arabidopsis thaliana suggest that the plasma membrane nitrate (NO3-) transporter NRT1.1 (CHL1), initially characterized as an influx carrier participating in the uptake of NO3- from soil solution (Tsay et al, 1993), also plays a role in NO3- signaling and sensing (Munõ s et al, 2004; Remans et al, 2006; Krouk et al, 2006, 2010; Ho et al, 2009; Wang et al, 2009). Studies have demonstrated AtNRT1.1 (AtNPF6.3), a member of the nitrate transporter 1/peptide transporter family for nitrate signaling in A. thaliana, as in addition to mediating uptake and transport of nitrate where AtNRT1.1 acts as a sensor to trigger the primary nitrate response (Ho et al, 2009). Monocots and eudicots differ in the number of their NRT1.1 gene, with monocotyledons typically having three to four members ofNRT1.1 and most eudicot species having only one NRT1.1 gene (Plett et al, 2010). Recently, Wen et al (2017) reported that two putative homologs of AtNRT1.1 (AtNPF6.3) in maize (Zea mays), ZmNPF6.4 and ZmNPF6.6, display distinct substrates affinity for nitrate and chloride, indicating a possible functional divergence among different NRT1.1 family members in mono- cotyledons (Wang et al, 2018). Five NRT2 genes have been identified in the rice genome (Cai et al, 2008; Feng et al, 2011). OsNRT2.1 and OsNRT2.2 share an identical coding region sequence with different 5ʹ - and 3ʹ -untranscribed regions and have high similarity to the NRT2 genes of other monocotyledons, while OsNRT2.3 and OsNRT2.4 are more closely related to Arabidopsis NRT2 genes (Tang et al, 2012). However, the biological functions and the possible divergence of the NRT1.1 and NRT2 nitrate transporter family have not been functionally characterized yet in rice.
For many higher plants like rice, nitrate is not only the main source of nitrogen but also acts as a major signal molecule controlling the plant metabolism and growth (Stitt, 1999). The signaling effect of NO3- is particularly strong on the development of lateral roots (LRs), which emerges post embryonically and determines the branching pattern of the root system (Forde, 2002; Malamy, 2005). For NO3- regulation in root system, NRT1.1 is crucial because it triggers a specific NO3- signaling pathway that stimulates lateral root (LR) growth in response to a localized supply of NO3- (Remans et al, 2006). As such, NRT1.1 plays an important role in the adaptive response of the plant to nitrogen limitation because it directs preferential growth of LRs in NO3- rich patches of the external medium. This review represents a major update on the N sensing and movement in rice plant, covering a wide range of genetic, molecular, physiological and developmental aspects of N nutrition. Furthermore, recent advances that will enable this fundamental research in model plants to improve N use efficiency in rice are considered.
Among the many different steps involved in the utilization of inorganic N by rice plants, those associated with the transport of nitrate and ammonium have received increasing attention over the past two decades. The first step of N acquisition by rice plant roots is active transport across the plasma membrane of root epidermal and cortical cells. It is now clear that many of the membrane transporter proteins have been identified at the molecular level in model species, and there is a general conservation of the related gene families between species, allowing rapid progress in research on different crops especially on rice plant. When N-deficient rice plants are supplied with a heterogeneous environment of nitrogen, they activate a set of morphological and physiological responses called N foraging condition (Bouguyon et al, 2015; O’ Brien et al, 2016). The most dramatic aspect of this response is enhancement of plant root and proliferation, particularly in NO3--rich zones and the local and systemic signaling regulation pathways of plant have been well reviewed (O’ Brien et al, 2016).
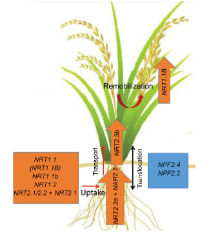
Under high NO3- conditions, the dual-affinity of nitrate transporter like NRT1.1 and the low-affinity transporter like NPF4.6 (NRT1.2/AIT1) in Arabidopsis (Wang et al, 2012; O’ Brien et al, 2016), the dual- affinity nitrate transporter MtNRT1.3 in medicago (Morè re-Le Paven et al, 2011), and the low affinity nitrate transporter OsNPF8.9 (OsNRT1.1, Os3g13274 or AF140606), the OsNRT1.1 allele OsNPF6.5 (OsNRT1.1B), OsNPF2.4 (OsNRT1.6), and a possible 6-transmembrane NO3- transporter OsNRT1.1b (AK066920) in rice (Hu et al, 2015; Xia et al, 2015; Fan et al, 2016a) are all shown to be involved in the uptake of NO3- by plant roots (Fig. 1). Under the limitation conditions of N, the NRT2 family plays an important role in NO3- acquisition from the soil environment (Morè re-Le Paven et al, 2011; Wang et al, 2012; Fan et al, 2016a; O’ Brien et al, 2016). In Arabidopsis, nitrate transporters like NRT2.1, NRT2.2, NRT2.4 and NRT2.5 require a partner protein, NAR2.1 (Kotur et al, 2012; Wang et al, 2012; Kotur and Glass, 2015; O’ Brien et al, 2016). Other NRT2s include CmNRT2.1 in chrysanthemum (Gu et al, 2016), TaNRT2.1 in wheat (Taulemesse et al, 2015), and OsNRT2.1, OsNRT2.2 and their partner protein OsNAR2.1 in rice (Feng et al, 2011; Yan et al, 2011; Liu et al, 2014), which function as NO3- influx in plant root as major components of high-affinity transport system (HATS) (Fig. 1). OsNRT1.1B is involved in nitrate utilization, and a single polymorphism in this gene contributes to the long-noted divergence in nitrogen use efficiency (NUE) between indica and japonica subspecies of Asian cultivated rice (Oryza sativa) (Hu et al, 2015). Similar expression and subcellular localization patterns, strongly suggest that OsNRT1.1B is the functional homolog of AtNRT1.1 for the functions in nitrate uptake, transport and signaling (Wang et al, 2018). Root NO3- efflux to the outer medium for net uptake of NO3- and can even exceed influx on exposure to various stresses such as drought, salinity, flooding, ozone, low nutrient availability pollutants, mechanical injury and pathogen.
The status of N in rice plant as well as local signals in roots trigger morphological changes in the whole system of root where NO3- acts locally to induce different morphological responses. This subject has been reviewed recently (le Deunff et al, 2016; Kiba and Krapp, 2016; O’ Brien et al, 2016). NRT1.1 and MtNRT1.3 can sense the external concentration of NO3- and alter their transport activity, and these proteins have been proposed as transceptors with the function of N-sensing in plants (Gojon et al, 2011; Morè re-Le Paven et al, 2011; Bouguyon et al, 2015). Auxin mediated NO3- signaling by NRT1.1 participates in the adaptive response of root architecture to the spatial heterogeneity of NO3- availability in rice plants (Mounier et al, 2014).
As N is one of the most important nutrients for plant growth and development, the sensing and signaling response of N are vital for plant growth, development and survive to the environment (Krouk et al, 2010). Plants have evolved several mechanisms for N uptake to support their survival in the changing environment. Nitrate and ammonium are the two main inorganic forms of N which exist in soils, of which nitrate is the major available form of N in the aerobic environments, whereas ammonium tends to be the major form in the flooded environment or acidic soil conditions (Ho and Tsay, 2010). Amino acids also account for a small portion of organic N that can be utilized by plants (Nä sholm et al, 2009; Moran-Zuloaga et al, 2015). In soil with low oxygen levels, ammonia is the primary nitrogen source, but toxicity is carefully controlled by plants with the transcription of ammonium transporters (AMTs) (Ho and Tsay, 2010). This metabolite and others including glutamate and glutamine have been shown to act as a signal of low nitrogen through regulation of nitrogen transporter gene transcription (Coruzzi and Zhou, 2001). NRT1.1, also known as CHL1, is the nitrate transceptor (transporter and receptor) found on the plasma membrane of plants (Ho and Tsay, 2010). This is both a high and low affinity transceptor that senses at varying concentrations of nitrate depending on its T101 residue phosphorylation (Ho and Tsay, 2010). It has been shown that nitrate can also act as just a signal for plants, since mutants unable to metabolize are still able to sense the ion (Coruzzi and Zhou, 2001). For example, many plants show the increase of nitrate- regulated genes in low nitrate conditions and consistent mRNA transcription of such genes in soil with high nitrate (Coruzzi and Zhou, 2001). This demonstrates the ability to sense nitrate soil concentrations without metabolic products of nitrate and still exhibit downstream genetic effects in plant bodies (Coruzzi and Zhou, 2001).
A few proteins have been identified in rice so far that take part in the activity of nitrate transport (Table 1). Among them, OsNRT2.1, OsNRT2.2 and OsNRT2.3a, belonging to the family of OsNRT2, are transcriptionally up-regulated by the supply of nitrate and require a partner protein OsNAR2.1 for nitrate uptake at varying concentrations (Table 1 and Fig. 1) (Tsay et al, 2007; Cai et al, 2008; Yan et al, 2011; Tang et al, 2012). OsNPF2.4 and OsNRT1.1b are two plasma membrane-localized nitrate transporters belonging to the nitrate transporter 1/peptide transporter (NPF) family (Hu et al, 2015; Xia et al, 2015), and also mediate the nitrate uptake from root and transport to shoot. In contrast to the NRT1 family transporters, the members of the NRT2 family transport nitrate with high affinity. The NAR2-like family genes are auxiliary partners for high affinity nitrate uptake (Kawachi et al, 2006; Okamoto et al, 2006; Orsel et al, 2006; Yong et al, 2010; Laugier et al, 2012). Disruption of AtNRT2.1 and AtNRT2.2 impairs the uptake of nitrate in both the inducible high-affinity transport system (IHATS) and the constitutive high-affinity transport system (CHATS) (Li et al, 2007). AtNRT2.4 is induced by low nitrate and mediates nitrate transport under the starvation of N in both rice shoots and roots (Kiba et al, 2012), whereas AtNRT2.5 plays an essential role in N-starved plants by ensuring efficient nitrate uptake and by participating in nitrate loading into the phloem during nitrate remobilization (Lezhneva et al, 2014; Kotur and Glass, 2015).
| Table 1 Different genes involved in N transport in rice plant. |
Three families of transporters such as NRT1, NRT2 (or NAR2/NRT2) and CLC have been identified for nitrate uptake and translocation in plants (Dechorgnat et al, 2011). Most members of NRT1 family have been characterized so far as low-affinity nitrate transporters and an exception is NRT1.1 (CHL1), which operates over both ranges. Some NRT2 members require a partner protein, NAR2, for nitrate transport at relatively low concentrations (Fig. 2). Expression of the NRTs is regulated by nitrate, N metabolites, N starvation, circadian rhythm, sucrose and pH (Krouk et al, 2010; Feng et al, 2011). Two nitrate-inducible kinases, CIPK8 and CIPK23, are either positive regulators for the low-affinity of NRT1.1 activity or negative regulators for the high-affinity phase of NRT1.1 (Ho et al, 2009). Such genetically distinct regulation of low and high affinity nitrate transport response indicates that there are likely to be differential regulators determining NUE at deficient and sufficient levels of N. There are five NRT2 family members in rice, each showing different affinities and regulation patterns by N supply form outside (Feng et al, 2011; Yan et al, 2011) (Fig. 2). Unlike its ortholog in Arabidopsis, the OsNAR2.1 accessory protein in rice interacts with three NRT2 transporters (NRT2.1, NRT2.2 and NRT2.3a) at both the protein levels and messenger RNA (mRNA) for nitrate uptake over both high and low N concentrations (Fig. 2). In addition, to compare the functions between mono and eudicotyledonous plants, it is important to understand the contribution and regulation of NRT family members to NUE for nitrate and ammonium preferring rice plants.
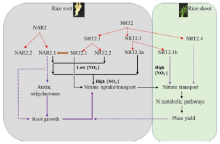 | Fig. 2. Schematic representation of proposed evolution and predicted functions for nitrate transporters (NAR2/NRT2) in rice. NAR2.1, NAR2.2, NRT2.1, NRT2.2 and NRT2.3a are expressed mainly in roots; NRT2.3b and NRT2.4 are expressed mainly in shoots (Feng et al, 2011; Yan et al, 2011). Both NRT2.1 and NRT2.2 associated with the nitrate transport (NAR2.1) in the high-affinity concentration range. NRT2.3a requires NAR2.1 for the nitrate transport function, and the protein has a 10-fold lower affinity for nitrate than NRT2.1 and NRT2.2. NAR2.1 can provide a switch, depending on the partner transporter, to enable a rapid response in nitrate uptake over the dynamic ranges of external concentrations of nitrate (Feng et al, 2011; Yan et al, 2011). In contrast, NRT2.3b can function in nitrate transport independently, mainly in the shoot, and its overexpression can greatly improve nitrogen use efficiency and grain yield in rice (Feng et al, 2011; Yan et al, 2011). The solid red arrows represent defined direct functions of the transporters in nitrate uptake and translocation; the dashed arrows represent presumed relationships based on the tissue localization of the genes in rice plant and functional expression in oocytes. The blue arrows indicate the proposed evolution of individual members of the NAR2 and NRT2 nitrate transporter families. Black arrows indicate the possible relationships between NAR2.1 and root growth and between the functions of NRT2 members and plant growth and development. |
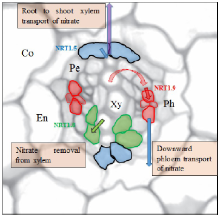
AtNPF6.3 is shown to function as an auxin efflux carrier in addition to its role for the transport of nitrate and is highly expressed in LR primordia, root cap and epidermis (Krouk et al, 2010), where shoot-ward auxin transport is required to mediate LR development (Xuan et al, 2016). Thus, auxin might act as a potential long-distance signal that mediates AtNPF6.3-dependent root development via its transport and accumulation. A dual-affinity nitrate transporter AtNPF6.3/AtCHL1/ AtNRT1.1 is demonstrated as a transporter and receptor for nitrate and regulates the emergence of LR upon nitrate starvation (Little et al, 2005; Ho et al, 2009; Krouk et al, 2010; Gojon et al, 2011; Xu et al, 2012; Krapp et al, 2014). AtNPF6.3 (CHL1/AtNRT1.1) is the first nitrate transporter identified to function not only the uptake of nitrate (Tsay et al, 1993) but also the translocation of nitrate from roots to shoots (Lé ran et al, 2013). It was also found to be a dual-affinity nitrate transporter, and the switch between the high- and low-affinity form is determined by the phosphorylation of Thr101 (Liu and Tsay, 2003). Presently, the crystal structure of AtNRT1.1 has been fixed by two independent groups (Parker and Newstead, 2014; Sun et al, 2014; Tsay, 2014), where phosphorylation of AtNRT1.1 can increase the structural flexibility and transport rates as well as phosphorylation-controlled dimerization switch which allows AtNRT1.1 for distinct affinity modes in rice plants. In addition to AtNRT1.1, AtNPF4.6 (AtNRT1.2) and AtNPF2.7 (NAXT1) are also involved in root nitrate uptake with influx and efflux activity, respectively (Huang et al, 1999; Segonzac et al, 2007; Kanno et al, 2012). AtNPF7.2 (AtNRT1.8), AtNPF7.3 (AtNRT1.5), AtNPF2.3 and AtNPF2.9 (AtNRT1.9) are major transporters for long-distance transport of nitrate, whereas AtNRT1.8 is predominantly expressed in xylem parenchyma cells within the vasculature and is important for nitrate unloading in xylem (Li J Y et al, 2010). Moreover, AtNRT1.5 is responsible for loading of nitrate in xylem, and mediate long-distance root to shoot transport (Lin et al, 2008) (Fig. 3). A similar expression pattern in roots was also observed for AtNPF2.3 which contributes to the secretion of nitrate into root xylem sap under salt stress (Taochy et al, 2015). Moreover, the transcription factor TCP20, miRNAs (e.g. miR169a and miR393), calcium and cytokinin have also been suggested as N-responsive long-distance signals in Arabidopsis (Rahayu et al, 2005; Vidal et al, 2010; Zhao et al, 2011; Guan et al, 2014; Riveras et al, 2015). It remains, however, unclear in Arabidopsis as well as in rice plants, whether they are moving throughout the plant to transmit the signals of N or act on their downstream signaling molecules to activate the scavenging responses.
Various external signals including rhizosphere pH, drought and soil micro-organisms (e.g. plant-growth- promoting rhizobacteria) in root-rhizosphere interaction zone can influence plant growth and development through the alteration in uptake and sensing of N in plants (Xu et al, 2012; He and Dijkstra, 2014; Fan et al, 2016b; Verbon and Liberman, 2016). Liu and von Wiré n (2017) and Undurraga et al (2017) document the role of both ammonium and nitrate as signaling molecules triggering a wide range of molecular, physiological and developmental responses in plants. Findings concerning the sensing mechanisms of these two ions, and the associated downstream signaling events are coming out at an unprecedented pace. Interestingly, both reviews highlighted the potential role of membrane transporters in ammonium or nitrate sensing, supporting the transceptor concept initially reported for the ammonium transporter (MEP2) in yeast (Holsbeeks et al, 2004). Lima et al (2010), Gojon et al (2011) and Bouguyon et al (2015) reported that the NPF6.3 (NRT1.1) nitrate transporter and the ammonium transporters (AMT1.1 and AMT1.3) serve as nitrate and ammonium sensors, respectively. Although the downstream signaling pathways still remain obscure at the molecular level for ammonium, a significant number of regulatory genes or secondary signals have recently been reported for nitrate (Undurraga et al, 2017), including NLP transcription factors (Marchive et al, 2013) and calcium (Riveras et al, 2015).
Plant nitrate transporters belonging to either nitrate and/or peptide transporter family (NPF/NRT1) or nitrate transporter 2 family (NRT2) are nitrate/proton symporters (Espen et al, 2004; Xu et al, 2012). Their transport activity of N is therefore found to be regulated by external (apoplastic) pH condition (Xia et al, 2015; Fan et al, 2016b). These observations indicate a possible regulatory role of external pH on nitrate sensing and signaling in rice plants. To reduce the fluctuation pH, plants may increase plasma membrane H+-ATPase activity to maintain the proton gradient during N uptake and assimilation (Zhu et al, 2009). For instance, the nitrate transceptor AtNPF6.3/AtCHL1/AtNRT1.1 displays the primary responses and transport activities of nitrate at low pH (about 5), but is less active as the pH is increased in culture medium to neutral condition (about 7) under both low and high conditions of nitrate (Ho et al, 2009). In rice, a low-affinity transporter like OsNPF2.4, is highly expressed in the root epidermis and may be involved in sensing and signaling of N. Recently, OsNRT2.3b, a splice variant of OsNRT2.3, has been found to play a critical role in sensing cytosol for pH changes and balancing the nitrate and ammonium uptake in rice plants (Fan et al, 2016b) (Fig. 1). This finding suggests that buffering of cytosolic pH can enhance the adaption of rice plant to rhizosphere pH by altering the activities of nitrate transporters.
Water availability also affects the conversion and concentration of nitrate and ammonium in rhizosphere zone of soil-plant interaction. To minimize the reduction in N uptake by drought stress, plants may activate specific genes or pathways to rescue the N uptake (He and Dijkstra, 2014). Overexpression of cytokinin synthesis gene isopentenyltrasnsferase(IPT), which is controlled by the promoter of a drought-inducible gene, is able to overcome the drought-caused inhibition of glutamine synthesis-dependent N assimilation and nitrate uptake in rice plants (Reguera et al, 2013). Interestingly, CHL1 or nodule inception like 7 protein(NLP7) mutation in Arabidopsis decreases the sensing, signaling and uptake activities of N, but enhances drought resistance in rice plants (Castaings et al, 2009; Marchive et al, 2013), suggesting that plants may also adapt their activity of N acquisition under drought stress for their survive to this environment.
Although the elucidation of sensing and signaling mechanisms of N now involves sophisticated functional genomics and system biology approaches, much remains to be done at the phenotypic level to unravel all effects of N for signaling, acquisition and utilization in plants to various environmental factors. The reviews by Lin and Tsay (2017) and Bloom and Rubio-Asensio (2017) are both excellent illustrations of this statement. Indeed, although it has been suspected for a long time that N compounds could be signal molecules controlling flowering and fruit setting in plants, Lin and Tsay (2017) stands as a rare example of a putative comprehensive model for explaining the effect of N availability on flowering time in Arabidopsis, and Bloom and Rubio-Asensio (2017) address the crucial question of the negative effect that the elevation of atmospheric concentration of CO2 is predicted to have on the status of N for most of the C3 plants.
In addition to the external signals in rice, the acquisition process of N is regulated by large number of genes at the levels of transcriptional, post- transcriptional and protein. The uptake of N by plants involves a complex gene regulatory networks, and nitrate-dependent signaling pathways in Arabidopsis have been intensively investigated and reviewed (O’ Brien et al, 2016; Bellegarde et al, 2017). Transcription factors (TFs) (e.g., MADS-box TF ANR1, LBD37, LBD38, NLP6, NLP7, GARP TF HRS1, TGA1, TGA4, SPL9 and AFB3) systemically regulate the transport, signaling and developmental responses of N-dependent phenomena (Zhang and Forde, 1998; Remans et al, 2006; Vidal et al, 2013; O’ Brien et al, 2016). In rice, orthologs of the above TFs (e.g. OsMADS, OsGATAs and OsGARP-G2-like TFs) are also found to be regulated under the starvation condition of N (Yang W Z et al, 2015), however, only few of them have been demonstrated in rice plants to regulate the developmental nitrate responses. For instance, OsMADS genes show diverse responses to nitrate supply in rice plants (Yu et al, 2014). Unlike AtANR1, which acts downstream of AtNPF6.3 (Remans et al, 2006), over-expression of a nitrate-inducible OsMADS25 gene significantly increases the expression of nitrate transporter genes and promotes accumulation of nitrate and LR and shoot growth, suggesting the positive effects on the uptake of nitrate via nitrate transporters (Yu et al, 2015). Interestingly, OsMADS genes are targeted by monocot specific Micro-RNA444s (Li et al, 2010) where OsmiR444a overexpression reduces the expression of OsMADS and root as well as shoot growth under nitrate supply condition in soil (Yan et al, 2014).
In plants, the transport activities of N also depend on phosphorylation triggered by external signals. For instance, the supply of high ammonium suppresses AtAMT1; 1 activity by phosphorylating Thr460 on the trans-activation domain in its C-terminus (Lanquar et al, 2009). Phosphorylation of AtAMTs at this site is controlled by CIPK23, which specifically interacts with AtAMT1; 1 and AtMT1; 2 in vivo, and negatively regulates the AtAMT1s-dependent uptake of N and growth responses of plant (Straub et al, 2017). The phosphorylation site Thr460 is also conserved in OsAMT1s (Yang S Y et al, 2015), however, its regulative role in OsAMT transport activity in rice plant has not been clarified yet. Recently, nitrogen limitation adaptation (NLA), a target of AtmiR827, is identified to interact with the nitrate transporter AtNRT1.7. The NLA negatively moderates AtNRT1.7 activities via the protein ubiquitination pathway, and thus regulates source-to-sink nitrate remobilization (Liu et al, 2017). In addition, the protein level of AtNPF6.3 is reduced in LR primordia under N supply, which is inconsistent with the changes of its local mRNA level, suggesting a translational regulation of AtNRT1.1 at the tissue-specific level of plant (Bouguyon et al, 2016). In rice plant, the OsNRT2.3b protein contains a pH-sensitive motif VYEAIHKI, and its transport activity is depending on intracellular pH and transmembrane potential, which are affected by both N uptake and assimilation (Fig. 2) (Fan et al, 2016b). Increasing pH sensitivity by over-expressing OsNRT2.3b stimulates the uptake and use efficiency of N and grain yield while genetic ablation of its function in sensing cytosolic pH changes represses the over-expression phenotype in rice plants (Fan et al, 2016b), which suggest a novel regulation of nitrate transport mechanism by cytosolic pH in rice plants.
A transcriptome analysis of root scavenging responses indicates that a long-distance systemic signaling triggered by nitrate sensing in rice plants is crucial to control the uptake of N (Ruffel et al, 2011). The bZIP family, which consists of DNA binding domain (BD) proteins rich in basic amino acids and adjacent to a leucine zipper dimerization domain, is one of the largest TF families in higher plants. There are 75 bZIPs in Arabidopsis (Jakoby et al, 2002) and 89 (Nijhawan et al, 2008) or 92 bZIPs in rice (Correa et al, 2008). HY5, a core bZIP TF expressed in the shoot, is required for light-regulated shoot development, and is found to regulate the uptake and assimilation of nitrate in root (Jonassen et al, 2008; Chen et al, 2016). In hy5 mutants, the uptake of N in the root tip is substantially decreased which can be rescued by shoot-derived or phloem-derived HY5 protein, implicating HY5 as a mobile signal that moves N through the phloem to activate root scavenging responses. Chip assays further suggest that HY5 binds directly to the promoter of the high-affinity transporter gene AtNRT2.1. This HY5-dependent induction occurs locally in the root to promote N uptake, and importantly, the binding activity relies on the carbon photo-assimilate (sucrose) levels in the shoot which highlight a systemic regulation of N uptake and C/N balance by a shoot- derived mobile molecule.
Evidence underlines the significance of small peptides in sensing and signaling homeostasis mechanism of N in plants (Okamoto et al, 2016). For instance, the N starvation-triggered C-terminally encoded peptide (CEP) is identified as ‘ hunger signal’ that regulates the development of root and nitrate uptake under the limited N condition (Tabata et al, 2014), whereas CLE (Clavata3/ESR-related)-peptide signal is found to repress nodule formation under N supply and proposed as a ‘ satiety signal’ for the availability of N (Okamoto et al, 2013). Both CLE and CEP peptide signals are secreted into the root vascular tissue, transmitted to the shoot, and perceived by their respective shoot- localized leucine-rich repeat receptor-like kinase (LRR-RLK) receptors HAR1 and CEPRs. Two phloem- specific polypeptides, CEP downstream1 (CEPD1) and CEPD2, are recently identified as CEP dependent downstream components. CEPDs are regulated by CEPRs and move root wards to activate AtNRT2.1 for N uptake (Ohkubo et al, 2017). Thus, CEL-HAR1, CEP-CEPR and their specific downstream signals may act as ‘ systemic signal modules’ to coordinate root and shoot development in response to the availability of N in soil.
In recent years, a remarkable increase in number of genes involved in the mechanisms for the utilization and assimilation of inorganic N has been shown in rice plants, especially for sensing, signaling and transporting mechanisms. In this century, a great challenge is to develop technologies leading to a sustainable agriculture. The use of chemical fertilizers cannot be eliminated without drastically decreasing food production. At the same time, there is an urgent need to lower the adverse environmental impacts of chemical fertilizers for sustainable agriculture. Different initiatives are in progress aiming to improve N nutrition and NUE in plants, such as the manipulation of plant N metabolism. Based on recent findings and views, we proposed a model for the mechanisms of how plants might transmit the local signal into a systemic signal for N uptake and plant foraging responses. However, many important clues for the sensing and signaling of N in rice plants are still missing. For instance, rice plants exhibit relative preferences for inorganic N sources in dry land (mainly nitrate) versus paddy field (ammonium). Rice grown in paddy soil acquires nitrate mostly at low nitrate level via high-affinity nitrate transporters that might be mediated by distinguished signaling pathways of N which are largely undetermined yet. Proton exchanges occur during the uptake and assimilation of N, and lead to alternation of both rhizosphere and cellular pH which could affect root growth and N uptake activity (Feng et al, 2011; Fan et al, 2016b; Pacheco-Villalobos et al, 2016). In addition, light signals, such as HY5 and a blue-light receptor Crypto- chrome 1 (CRY1), are found to mediate N-dependent root growth and flowering in Arabidopsis (Chen et al, 2016; Yuan et al, 2016), but it remains unclear for rice plant whether they are active in a similar pattern. Recently, the OsCEP peptides have been identified in rice, which are shown to negatively regulate rice growth (Sui et al, 2016), however, it is not clear whether OsCEPs involve N-dependent sensing and signaling pathways.
In conclusion, there is an immediate need for future in-depth studies that should concentrate on the role of mobile molecules such as peptides and micro-RNAs in the N-dependent signaling pathways, and on system biology approaches to explore novel sensing and signaling components that are involved in N assimilation in crop species especially in rice. In rice, the signaling network of N has gained new levels of complexity during very recent years and is as yet far from understood. Indeed, many regulatory elements are probably still missing in the response pathway of nitrate, and the connections which exist between this signaling pathway and others responding to nutrients, stress, or hormones remain to be established. In the future, global approaches will probably help to combine the traditional forward genetics together with the newer genomics and cell-specific techniques and to build a coherent and comprehensive plant N sensing and signaling network in rice plants.
The author is grateful for financial support from the Research and Training Center (RTC) at Patuakhali Science and Technology University (PSTU), Dumki, Patuakhali-8602, Bangladesh (Grant No. 4829) and also for Research Collaboration Fund provided by the University Grants Commission, Bangladesh.
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
|
| [56] |
|
| [57] |
|
| [58] |
|
| [59] |
|
| [60] |
|
| [61] |
|
| [62] |
|
| [63] |
|
| [64] |
|
| [65] |
|
| [66] |
|
| [67] |
|
| [68] |
|
| [69] |
|
| [70] |
|
| [71] |
|
| [72] |
|
| [73] |
|
| [74] |
|
| [75] |
|
| [76] |
|
| [77] |
|
| [78] |
|
| [79] |
|
| [80] |
|
| [81] |
|
| [82] |
|
| [83] |
|
| [84] |
|
| [85] |
|
| [86] |
|
| [87] |
|
| [88] |
|
| [89] |
|
| [90] |
|
| [91] |
|
| [92] |
|
| [93] |
|
| [94] |
|
| [95] |
|
| [96] |
|
| [97] |
|
| [98] |
|
| [99] |
|
| [100] |
|
| [101] |
|
| [102] |
|
| [103] |
|
| [104] |
|
| [105] |
|
| [106] |
|
| [107] |
|
| [108] |
|
| [109] |
|
| [110] |
|
| [111] |
|
| [112] |
|
| [113] |
|
| [114] |
|
| [115] |
|
| [116] |
|
| [117] |
|
| [118] |
|
| [119] |
|
| [120] |
|
| [121] |
|
| [122] |
|