
Rice Science ›› 2015, Vol. 22 ›› Issue (6): 264-274.DOI: 10.1016/S1672-6308(14)60306-1
收稿日期:2015-04-06
接受日期:2015-05-08
出版日期:2015-06-06
发布日期:2015-09-15
. [J]. Rice Science, 2015, 22(6): 264-274.
| Sampling stage | Treatment | Organic matter | Total N | Alkali-hydrolyzable | Olsen-P | Available K | pH |
|---|---|---|---|---|---|---|---|
| (g/kg) | (g/kg) | N (g/kg) | (g/kg) | (g/kg) | |||
| Before sowing | A | 13.71 | 1.11 | 0.06 | 0.04 | 0.1 | 8.4 |
| B | 7.85 | 0.57 | 0.04 | 0.01 | 0.07 | 8.6 | |
| C | 8.2 | 0.62 | 0.04 | 0.01 | 0.07 | 8.6 | |
| Elongation stage (7 d after the first fertilizer application) | A | - | 1.12 | 0.08 | 0.04 | - | - |
| B | - | 0.55 | 0.04 | 0.01 | - | - | |
| C | - | 0.56 | 0.04 | 0.01 | - | - | |
| Before booting stage (before the second fertilizer application) | A | - | 0.96 | 0.07 | 0.03 | - | - |
| B | - | 0.51 | 0.04 | 0.01 | - | - | |
| C | - | 0.53 | 0.04 | 0.01 | - | - | |
| Booting stage (7 d after the second fertilizer application) | A | - | 0.95 | 0.06 | 0.03 | - | - |
| B | - | 0.55 | 0.05 | 0.01 | - | - | |
| C | - | 0.48 | 0.04 | 0.01 | - | - | |
| After harvest | A | 13.35 | 1.02 | 0.06 | 0.03 | 0.08 | 8.2 |
| B | 7.42 | 0.53 | 0.03 | 0.01 | 0.06 | 8.2 | |
| C | 7.05 | 0.54 | 0.03 | 0.01 | 0.06 | 8.2 |
Table 1 Soil organic matter and nitrogen (N), phosphorus (P) and potassium (K) contents of trial area for different treatments in Langfang, Hebei Province, China.
| Sampling stage | Treatment | Organic matter | Total N | Alkali-hydrolyzable | Olsen-P | Available K | pH |
|---|---|---|---|---|---|---|---|
| (g/kg) | (g/kg) | N (g/kg) | (g/kg) | (g/kg) | |||
| Before sowing | A | 13.71 | 1.11 | 0.06 | 0.04 | 0.1 | 8.4 |
| B | 7.85 | 0.57 | 0.04 | 0.01 | 0.07 | 8.6 | |
| C | 8.2 | 0.62 | 0.04 | 0.01 | 0.07 | 8.6 | |
| Elongation stage (7 d after the first fertilizer application) | A | - | 1.12 | 0.08 | 0.04 | - | - |
| B | - | 0.55 | 0.04 | 0.01 | - | - | |
| C | - | 0.56 | 0.04 | 0.01 | - | - | |
| Before booting stage (before the second fertilizer application) | A | - | 0.96 | 0.07 | 0.03 | - | - |
| B | - | 0.51 | 0.04 | 0.01 | - | - | |
| C | - | 0.53 | 0.04 | 0.01 | - | - | |
| Booting stage (7 d after the second fertilizer application) | A | - | 0.95 | 0.06 | 0.03 | - | - |
| B | - | 0.55 | 0.05 | 0.01 | - | - | |
| C | - | 0.48 | 0.04 | 0.01 | - | - | |
| After harvest | A | 13.35 | 1.02 | 0.06 | 0.03 | 0.08 | 8.2 |
| B | 7.42 | 0.53 | 0.03 | 0.01 | 0.06 | 8.2 | |
| C | 7.05 | 0.54 | 0.03 | 0.01 | 0.06 | 8.2 |
| Material | Treatment | RRL (cm) | RRW (g) | RSW (g) | RWRSR | RTW (g) |
|---|---|---|---|---|---|---|
| Shuhui 527 | ||||||
| Mean ± SD | E-D | 1.8 ± 2.6 a | 0.015 ± 0.003 b | 0.048 ± 0.018 a | 0.039 ± 0.027 b | 0.062 ± 0.021 a |
| F-D | 4.0 ± 0.3 a | 0.026 ± 0.000 a | 0.030 ± 0.009 a | 0.207 ± 0.035 a | 0.056 ± 0.009 a | |
| Minghui 86 | ||||||
| Mean ± SD | E-D | 0.1 ± 1.0 a | 0.006 ± 0.008 a | 0.011 ± 0.015 a | 0.011 ± 0.067 b | -0.005 ± 0.062 a |
| F-D | 0.6 ± 0.9 a | 0.008 ± 0.009 a | -0.005 ± 0.023 a | 0.160 ± 0.086 a | 0.003 ± 0.027 a | |
| Yetuozai (1) | ||||||
| Mean ± SD | E-D | 0.5 ± 1.8 a | 0.003 ± 0.012 a | 0.006 ± 0.038 a | 0.021 ± 0.082 a | 0.010 ± 0.049 a |
| F-D | -0.1 ± 2.8 a | 0.009 ± 0.009 a | 0.001 ± 0.022 a | 0.107 ± 0.086 a | 0.009 ± 0.029 a | |
| Yetuozai (2) | ||||||
| Mean ± SD | E-D | 0.1 ± 1.1 b | 0.001 ± 0.007 a | -0.001 ± 0.008 a | 0.013 ± 0.071 a | -0.002 ± 0.015 a |
| F-D | 1.8 ± 1.6 a | 0.001 ± 0.007 a | -0.021 ± 0.020 b | 0.135 ± 0.144 a | -0.020 ± 0.022 a | |
| Shuhui 527/Yetuozai population (SY) | ||||||
| Mean ± SD | E-D | 0.3 ± 0.9 a | 0.005 ± 0.017 a | 0.001 ± 0.033 a | 0.023 ± 0.042 a | 0.008 ± 0.045 a |
| F-D | 0.6 ± 1.1 a | 0.007 ± 0.010 a | -0.015 ± 0.035 a | 0.102 ± 0.072 a | -0.007 ± 0.042 a | |
| Range | E-D | -2.1-2.9 | -0.037-0.097 | -0.153-0.059 | -0.102-0.131 | -0.180-0.125 |
| F-D | -2.8-3.1 | -0.015-0.040 | -0.116-0.073 | -0.084-0.406 | -0.131-0.093 | |
| Minghui 86/Yetuozai population (MY) | ||||||
| Mean ± SD | E-D | 0.2 ± 1.2 b | 0.001 ± 0.008 a | -0.007 ± 0.019 a | 0.026 ± 0.044 b | -0.006 ± 0.025 a |
| F-D | 0.9 ± 1.0 a | 0.004 ± 0.009 a | -0.027 ± 0.019 b | 0.154 ± 0.112 a | -0.023 ± 0.026 b | |
| Range | E-D | -2.4-1.6 | -0.014-0.014 | -0.043-0.044 | -0.070-0.127 | -0.057-0.057 |
| F-D | -1.3-3.0 | -0.011-0.030 | -0.065-0.024 | 0.054-0.839 | -0.064-0.054 | |
Table 3 Phenotypic performances of measured traits for introgression lines and their parents under nutrient solution culture conditions in Beijing, China.
| Material | Treatment | RRL (cm) | RRW (g) | RSW (g) | RWRSR | RTW (g) |
|---|---|---|---|---|---|---|
| Shuhui 527 | ||||||
| Mean ± SD | E-D | 1.8 ± 2.6 a | 0.015 ± 0.003 b | 0.048 ± 0.018 a | 0.039 ± 0.027 b | 0.062 ± 0.021 a |
| F-D | 4.0 ± 0.3 a | 0.026 ± 0.000 a | 0.030 ± 0.009 a | 0.207 ± 0.035 a | 0.056 ± 0.009 a | |
| Minghui 86 | ||||||
| Mean ± SD | E-D | 0.1 ± 1.0 a | 0.006 ± 0.008 a | 0.011 ± 0.015 a | 0.011 ± 0.067 b | -0.005 ± 0.062 a |
| F-D | 0.6 ± 0.9 a | 0.008 ± 0.009 a | -0.005 ± 0.023 a | 0.160 ± 0.086 a | 0.003 ± 0.027 a | |
| Yetuozai (1) | ||||||
| Mean ± SD | E-D | 0.5 ± 1.8 a | 0.003 ± 0.012 a | 0.006 ± 0.038 a | 0.021 ± 0.082 a | 0.010 ± 0.049 a |
| F-D | -0.1 ± 2.8 a | 0.009 ± 0.009 a | 0.001 ± 0.022 a | 0.107 ± 0.086 a | 0.009 ± 0.029 a | |
| Yetuozai (2) | ||||||
| Mean ± SD | E-D | 0.1 ± 1.1 b | 0.001 ± 0.007 a | -0.001 ± 0.008 a | 0.013 ± 0.071 a | -0.002 ± 0.015 a |
| F-D | 1.8 ± 1.6 a | 0.001 ± 0.007 a | -0.021 ± 0.020 b | 0.135 ± 0.144 a | -0.020 ± 0.022 a | |
| Shuhui 527/Yetuozai population (SY) | ||||||
| Mean ± SD | E-D | 0.3 ± 0.9 a | 0.005 ± 0.017 a | 0.001 ± 0.033 a | 0.023 ± 0.042 a | 0.008 ± 0.045 a |
| F-D | 0.6 ± 1.1 a | 0.007 ± 0.010 a | -0.015 ± 0.035 a | 0.102 ± 0.072 a | -0.007 ± 0.042 a | |
| Range | E-D | -2.1-2.9 | -0.037-0.097 | -0.153-0.059 | -0.102-0.131 | -0.180-0.125 |
| F-D | -2.8-3.1 | -0.015-0.040 | -0.116-0.073 | -0.084-0.406 | -0.131-0.093 | |
| Minghui 86/Yetuozai population (MY) | ||||||
| Mean ± SD | E-D | 0.2 ± 1.2 b | 0.001 ± 0.008 a | -0.007 ± 0.019 a | 0.026 ± 0.044 b | -0.006 ± 0.025 a |
| F-D | 0.9 ± 1.0 a | 0.004 ± 0.009 a | -0.027 ± 0.019 b | 0.154 ± 0.112 a | -0.023 ± 0.026 b | |
| Range | E-D | -2.4-1.6 | -0.014-0.014 | -0.043-0.044 | -0.070-0.127 | -0.057-0.057 |
| F-D | -1.3-3.0 | -0.011-0.030 | -0.065-0.024 | 0.054-0.839 | -0.064-0.054 | |
| Treatment | Population | Trait | QTL | Marker | Chromosome | Position (cM) | Bin | Probability | R2 (%) | Additive |
|---|---|---|---|---|---|---|---|---|---|---|
| A | SY | PN | qPN5.7 | RM26 | 5 | 118.8 | 5.7 | 0.017 | 10.2 | 0.47 |
| A | SY | PN | qPN11.3 | RM536 | 11 | 55.1 | 11.3 | 0.005 | 13 | 0.29 |
| A | MY | PN | qPN5.3 | RM169 | 5 | 58 | 5.3 | 0.032 | 8.5 | -0.3 |
| A | SY | FGP | qFGP11.6 | RM254 | 11 | 110 | 11.6 | 0.017 | 9.7 | -8 |
| A | SY | SNP | qSNP6.6 | RM5463 | 6 | 124.4 | 6.6 | 0.016 | 9.8 | 10.48 |
| A | SY | SNP | qSNP11.6 | RM254 | 11 | 110 | 11.6 | 0.018 | 9.5 | -9.11 |
| A | SY | SF | qSF5.5 | RM164 | 5 | 78.7 | 5.5 | 0.015 | 10 | 1.52 |
| A | MY | SF | qSF4.1 | RM335 | 4 | 22 | 4.1 | 0.014 | 11 | -2.53 |
| A | MY | SF | qSF12.1 | RM20A | 12 | 3.2 | 12.1 | 0.006 | 13.8 | -4.51 |
| A | SY | TGW | qTGW4.1 | RM335 | 4 | 22 | 4.1 | 0.025 | 8.6 | -1.05 |
| A | SY | TGW | qTGW5.5 | RM164 | 5 | 78.7 | 5.5 | 0.04 | 7.2 | 0.63 |
| A | SY | TGW | qTGW6.6 | RM5463 | 6 | 124.4 | 6.6 | 0.008 | 11.9 | -1 |
| A | SY | TGW | qTGW9.3 | RM219 | 9 | 11.7 | 9.3 | 0.017 | 9.5 | -0.83 |
| A | MY | TGW | qTGW1.10 | RM200 | 1 | 143.2 | 1.1 | 0.031 | 8.7 | 0.55 |
| A | MY | TGW | qTGW3.12 | RM514 | 3 | 216.4 | 3.12 | 0.01 | 12 | 0.64 |
| A | SY | GY | qGY6.5 | RM541 | 6 | 76 | 6.5 | 0.009 | 11 | 1.15 |
| A | SY | GY | qGY11.6 | RM254 | 11 | 110 | 11.6 | 0.018 | 9.5 | -1.06 |
| B | SY | PN | qPN1.7 | RM9 | 1 | 92.4 | 1.7 | 0.047 | 6.8 | 0.32 |
| B | SY | PN | qPN11.3 | RM536 | 11 | 55.1 | 11.3 | 0.003 | 14.4 | 0.33 |
| B | SY | PN | qPN12.2 | RM155 | 12 | 20.9 | 12.2 | 0.004 | 14.4 | 0.25 |
| B | SY | FGP | qFGP11.6 | RM254 | 11 | 110 | 11.6 | 0.007 | 12.2 | -8.56 |
| B | MY | FGP | qFGP12.4 | RM511 | 12 | 59.8 | 12.4 | 0.007 | 13.3 | 14.57 |
| B | SY | SNP | qSNP11.6 | RM254 | 11 | 110 | 11.6 | 0.006 | 12.7 | -9.26 |
| B | MY | SNP | qSNP12.4 | RM511 | 12 | 59.8 | 12.4 | 0.003 | 16.1 | 18.81 |
| B | SY | SF | qSF1.7 | RM9 | 1 | 92.4 | 1.7 | 0.022 | 8.9 | -2.3 |
| B | SY | TGW | qTGW4.1 | RM335 | 4 | 22 | 4.1 | 0.005 | 12.8 | -1.32 |
| B | SY | TGW | qTGW6.6 | RM5463 | 6 | 124.4 | 6.6 | 0.002 | 15.1 | -1.17 |
| B | SY | TGW | qTGW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.046 | 6.7 | 0.9 |
| B | SY | TGW | qTGW9.3 | RM219 | 9 | 11.7 | 9.3 | 0.014 | 10 | -0.88 |
| B | MY | TGW | qTGW1.10 | RM200 | 1 | 143.2 | 1.1 | 0.002 | 16.5 | 0.69 |
| B | SY | GY | qGY6.5 | RM541 | 6 | 76 | 6.5 | 0.024 | 8.5 | 0.88 |
| B | SY | GY | qGY11.6 | RM254 | 11 | 110 | 11.6 | 0.013 | 10.4 | -0.98 |
| C | SY | PN | qPN1.2 | RM283 | 1 | 31.4 | 1.2 | 0.001 | 16.5 | -0.42 |
| C | SY | PN | qPN1.7 | RM9 | 1 | 92.4 | 1.7 | 0.005 | 13 | 0.54 |
| C | SY | PN | qPN5.7 | RM26 | 5 | 118.8 | 5.7 | 0.007 | 12.6 | 0.65 |
| C | SY | PN | qPN6.1 | RM589 | 6 | 3.2 | 6.1 | 0.008 | 11.4 | 0.33 |
| C | SY | PN | qPN11.3 | RM536 | 11 | 55.1 | 11.3 | 0.007 | 11.8 | 0.35 |
| C | MY | PN | qPN5.3 | RM169 | 5 | 58 | 5.3 | 0.015 | 11 | -0.44 |
| C | SY | SNP | qSNP6.6 | RM5463 | 6 | 124.4 | 6.6 | 0.02 | 9.2 | 9.87 |
| C | SY | SNP | qSNP11.6 | RM254 | 11 | 110 | 11.6 | 0.039 | 7.3 | -7.75 |
| C | SY | SF | qSF1.7 | RM9 | 1 | 92.4 | 1.7 | 0.003 | 14.9 | -3.42 |
| C | SY | SF | qSF5.5 | RM164 | 5 | 78.7 | 5.5 | 0.018 | 9.4 | 1.62 |
| C | MY | SF | qSF4.1 | RM335 | 4 | 22 | 4.1 | 0.043 | 7.6 | -2.06 |
| C | SY | TGW | qTGW4.1 | RM335 | 4 | 22 | 4.1 | 0.007 | 12.3 | -1.08 |
| C | SY | TGW | qTGW5.5 | RM164 | 5 | 78.7 | 5.5 | 0.004 | 13.7 | 0.74 |
| C | SY | TGW | qTGW6.6 | RM5463 | 6 | 124.4 | 6.6 | 0.004 | 13.7 | -0.92 |
| C | SY | TGW | qTGW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.032 | 7.7 | 0.8 |
| C | SY | TGW | qTGW9.3 | RM219 | 9 | 11.7 | 9.3 | 0.013 | 10.1 | -0.74 |
| C | MY | GY | qGY1.10 | RM200 | 1 | 143.2 | 1.1 | 0.003 | 15.8 | 1.42 |
Table 4 Identification of QTLs for yield traits in two populations under three treatments in Langfang, Hebei Province, China.
| Treatment | Population | Trait | QTL | Marker | Chromosome | Position (cM) | Bin | Probability | R2 (%) | Additive |
|---|---|---|---|---|---|---|---|---|---|---|
| A | SY | PN | qPN5.7 | RM26 | 5 | 118.8 | 5.7 | 0.017 | 10.2 | 0.47 |
| A | SY | PN | qPN11.3 | RM536 | 11 | 55.1 | 11.3 | 0.005 | 13 | 0.29 |
| A | MY | PN | qPN5.3 | RM169 | 5 | 58 | 5.3 | 0.032 | 8.5 | -0.3 |
| A | SY | FGP | qFGP11.6 | RM254 | 11 | 110 | 11.6 | 0.017 | 9.7 | -8 |
| A | SY | SNP | qSNP6.6 | RM5463 | 6 | 124.4 | 6.6 | 0.016 | 9.8 | 10.48 |
| A | SY | SNP | qSNP11.6 | RM254 | 11 | 110 | 11.6 | 0.018 | 9.5 | -9.11 |
| A | SY | SF | qSF5.5 | RM164 | 5 | 78.7 | 5.5 | 0.015 | 10 | 1.52 |
| A | MY | SF | qSF4.1 | RM335 | 4 | 22 | 4.1 | 0.014 | 11 | -2.53 |
| A | MY | SF | qSF12.1 | RM20A | 12 | 3.2 | 12.1 | 0.006 | 13.8 | -4.51 |
| A | SY | TGW | qTGW4.1 | RM335 | 4 | 22 | 4.1 | 0.025 | 8.6 | -1.05 |
| A | SY | TGW | qTGW5.5 | RM164 | 5 | 78.7 | 5.5 | 0.04 | 7.2 | 0.63 |
| A | SY | TGW | qTGW6.6 | RM5463 | 6 | 124.4 | 6.6 | 0.008 | 11.9 | -1 |
| A | SY | TGW | qTGW9.3 | RM219 | 9 | 11.7 | 9.3 | 0.017 | 9.5 | -0.83 |
| A | MY | TGW | qTGW1.10 | RM200 | 1 | 143.2 | 1.1 | 0.031 | 8.7 | 0.55 |
| A | MY | TGW | qTGW3.12 | RM514 | 3 | 216.4 | 3.12 | 0.01 | 12 | 0.64 |
| A | SY | GY | qGY6.5 | RM541 | 6 | 76 | 6.5 | 0.009 | 11 | 1.15 |
| A | SY | GY | qGY11.6 | RM254 | 11 | 110 | 11.6 | 0.018 | 9.5 | -1.06 |
| B | SY | PN | qPN1.7 | RM9 | 1 | 92.4 | 1.7 | 0.047 | 6.8 | 0.32 |
| B | SY | PN | qPN11.3 | RM536 | 11 | 55.1 | 11.3 | 0.003 | 14.4 | 0.33 |
| B | SY | PN | qPN12.2 | RM155 | 12 | 20.9 | 12.2 | 0.004 | 14.4 | 0.25 |
| B | SY | FGP | qFGP11.6 | RM254 | 11 | 110 | 11.6 | 0.007 | 12.2 | -8.56 |
| B | MY | FGP | qFGP12.4 | RM511 | 12 | 59.8 | 12.4 | 0.007 | 13.3 | 14.57 |
| B | SY | SNP | qSNP11.6 | RM254 | 11 | 110 | 11.6 | 0.006 | 12.7 | -9.26 |
| B | MY | SNP | qSNP12.4 | RM511 | 12 | 59.8 | 12.4 | 0.003 | 16.1 | 18.81 |
| B | SY | SF | qSF1.7 | RM9 | 1 | 92.4 | 1.7 | 0.022 | 8.9 | -2.3 |
| B | SY | TGW | qTGW4.1 | RM335 | 4 | 22 | 4.1 | 0.005 | 12.8 | -1.32 |
| B | SY | TGW | qTGW6.6 | RM5463 | 6 | 124.4 | 6.6 | 0.002 | 15.1 | -1.17 |
| B | SY | TGW | qTGW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.046 | 6.7 | 0.9 |
| B | SY | TGW | qTGW9.3 | RM219 | 9 | 11.7 | 9.3 | 0.014 | 10 | -0.88 |
| B | MY | TGW | qTGW1.10 | RM200 | 1 | 143.2 | 1.1 | 0.002 | 16.5 | 0.69 |
| B | SY | GY | qGY6.5 | RM541 | 6 | 76 | 6.5 | 0.024 | 8.5 | 0.88 |
| B | SY | GY | qGY11.6 | RM254 | 11 | 110 | 11.6 | 0.013 | 10.4 | -0.98 |
| C | SY | PN | qPN1.2 | RM283 | 1 | 31.4 | 1.2 | 0.001 | 16.5 | -0.42 |
| C | SY | PN | qPN1.7 | RM9 | 1 | 92.4 | 1.7 | 0.005 | 13 | 0.54 |
| C | SY | PN | qPN5.7 | RM26 | 5 | 118.8 | 5.7 | 0.007 | 12.6 | 0.65 |
| C | SY | PN | qPN6.1 | RM589 | 6 | 3.2 | 6.1 | 0.008 | 11.4 | 0.33 |
| C | SY | PN | qPN11.3 | RM536 | 11 | 55.1 | 11.3 | 0.007 | 11.8 | 0.35 |
| C | MY | PN | qPN5.3 | RM169 | 5 | 58 | 5.3 | 0.015 | 11 | -0.44 |
| C | SY | SNP | qSNP6.6 | RM5463 | 6 | 124.4 | 6.6 | 0.02 | 9.2 | 9.87 |
| C | SY | SNP | qSNP11.6 | RM254 | 11 | 110 | 11.6 | 0.039 | 7.3 | -7.75 |
| C | SY | SF | qSF1.7 | RM9 | 1 | 92.4 | 1.7 | 0.003 | 14.9 | -3.42 |
| C | SY | SF | qSF5.5 | RM164 | 5 | 78.7 | 5.5 | 0.018 | 9.4 | 1.62 |
| C | MY | SF | qSF4.1 | RM335 | 4 | 22 | 4.1 | 0.043 | 7.6 | -2.06 |
| C | SY | TGW | qTGW4.1 | RM335 | 4 | 22 | 4.1 | 0.007 | 12.3 | -1.08 |
| C | SY | TGW | qTGW5.5 | RM164 | 5 | 78.7 | 5.5 | 0.004 | 13.7 | 0.74 |
| C | SY | TGW | qTGW6.6 | RM5463 | 6 | 124.4 | 6.6 | 0.004 | 13.7 | -0.92 |
| C | SY | TGW | qTGW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.032 | 7.7 | 0.8 |
| C | SY | TGW | qTGW9.3 | RM219 | 9 | 11.7 | 9.3 | 0.013 | 10.1 | -0.74 |
| C | MY | GY | qGY1.10 | RM200 | 1 | 143.2 | 1.1 | 0.003 | 15.8 | 1.42 |
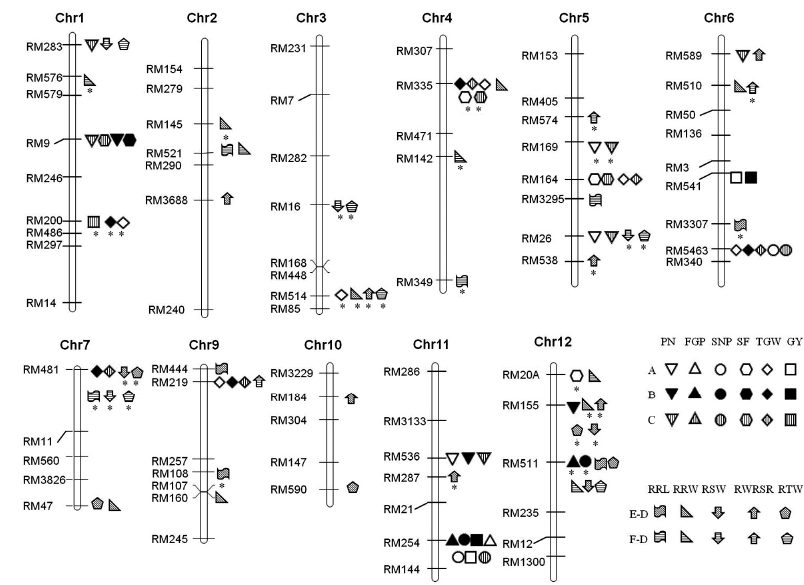
Fig. 1. Identified QTLs for measured traits in two introgressed line populations under two environments.(A, Normal fertilization treatment in normal soil; B, Normal fertilization treatment in barren soil; C, Low-phosphorus stress in barren soil; PN, Panicle number per plant; FGP, Filled grain number per panicle; SNP, Spikelet number per panicle; SF, Spikelet fertility; TGW, 1000-grain weight; GY, Grain yield per plant; RRL, Relative root length; RRW, Relative root dry weight; RSW, Relative shoot dry weight; RTW, Relative total dry weight; RWRSR, Relative root-shoot ratio of dry weight; D, Control; E, Nutrient solution with 1/3 phosphorus of the control; F, Nutrient solution without phosphorus; Chr, Chromosome. * below the icon represents the QTL identified in Minghui 86/Yetuozai population, otherwise, in Shuhui 527/Yetuozai population.)
| Treatment | Population | Trait | QTL | Marker | Chromosome | Position (cM) | Bin | Probability | R2 (%) | Additive |
|---|---|---|---|---|---|---|---|---|---|---|
| E-D | SY | RRL | qRRL9.3 | RM444 | 9 | 3.3 | 9.3 | 0.014 | 11.5 | 0.397 |
| E-D | SY | RRL | qRRL12.4 | RM511 | 12 | 59.8 | 12.4 | 0.009 | 13 | 0.289 |
| E-D | MY | RRL | qRRL6.6 | RM3307 | 6 | 113.4 | 6.6 | 0.007 | 14.4 | -0.403 |
| E-D | MY | RRL | qRRL9.7 | RM108 | 9 | 73.3 | 9.7 | 0.034 | 11.4 | 0.353 |
| E-D | SY | RRW | qRRW4.1 | RM335 | 4 | 21.5 | 4.1 | 0.019 | 10.9 | 0.009 |
| E-D | SY | RRW | qRRW6.3 | RM510 | 6 | 20.8 | 6.3 | 0.035 | 8.8 | -0.007 |
| E-D | SY | RRW | qRRW7.7 | RM47 | 7 | 90 | 7.7 | 0.036 | 8.8 | -0.009 |
| E-D | SY | RRW | qRRW9.7 | RM160 | 9 | 82.4 | 9.7 | 0.03 | 9.9 | 0.006 |
| E-D | MY | RRW | qRRW2.3 | RM145 | 2 | 49.8 | 2.3 | 0.028 | 9.6 | 0.002 |
| E-D | MY | RRW | qRRW3.12 | RM514 | 3 | 216.4 | 3.12 | 0.049 | 7.9 | -0.002 |
| E-D | MY | RRW | qRRW12.2 | RM155 | 12 | 20.9 | 12.2 | 0.016 | 11.5 | 0.003 |
| E-D | SY | RSW | qRSW12.4 | RM511 | 12 | 59.8 | 12.4 | 0.037 | 8.4 | -0.01 |
| E-D | MY | RSW | qRSW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.032 | 9.2 | -0.004 |
| E-D | MY | RSW | qRSW12.2 | RM155 | 12 | 20.9 | 12.2 | 0.04 | 8.5 | 0.007 |
| E-D | SY | RTW | qRTW7.7 | RM47 | 7 | 90 | 7.7 | 0.039 | 8.7 | -0.024 |
| E-D | SY | RTW | qRTW12.4 | RM511 | 12 | 59.8 | 12.4 | 0.024 | 10.2 | -0.014 |
| E-D | MY | RTW | qRTW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.038 | 8.7 | -0.006 |
| E-D | MY | RTW | qRTW10.6 | RM590 | 10 | 117.2 | 10.6 | 0.046 | 8 | -0.01 |
| E-D | MY | RTW | qRTW12.2 | RM155 | 12 | 20.9 | 12.2 | 0.035 | 9 | 0.01 |
| E-D | SY | RWRSR | qRWRSR2.5 | RM3688 | 2 | 88.2 | 2.5 | 0.02 | 10.8 | -0.019 |
| E-D | SY | RWRSR | qRWRSR6.1 | RM589 | 6 | 3.2 | 6.1 | 0.022 | 10.3 | -0.017 |
| E-D | SY | RWRSR | qRWRSR10.5 | RM184 | 10 | 58 | 10.5 | 0.008 | 13.4 | 0.015 |
| E-D | MY | RWRSR | qRWRSR3.12 | RM514 | 3 | 216.4 | 3.12 | 0.019 | 11 | -0.015 |
| E-D | MY | RWRSR | qRWRSR5.8 | RM538 | 5 | 132.7 | 5.8 | 0.046 | 8 | 0.021 |
| E-D | MY | RWRSR | qRWRSR11.4 | RM287 | 11 | 68.6 | 11.4 | 0.048 | 7.9 | 0.012 |
| E-D | MY | RWRSR | qRWRSR12.2 | RM155 | 12 | 20.9 | 12.2 | 0.005 | 15 | 0.007 |
| F-D | SY | RRL | qRRL2.4 | RM521 | 2 | 62.2 | 2.4 | 0.005 | 14.5 | 0.392 |
| F-D | SY | RRL | qRRL5.6 | RM3295 | 5 | 91.2 | 5.6 | 0.003 | 16.6 | -0.489 |
| F-D | MY | RRL | qRRL4.8 | RM349 | 4 | 146.8 | 4.8 | 0.006 | 15.1 | 0.835 |
| F-D | MY | RRL | qRRL7.1 | RM481 | 7 | 3.2 | 7.1 | 0.015 | 12.1 | 0.166 |
| F-D | SY | RRW | qRRW2.4 | RM290 | 2 | 66 | 2.4 | 0.047 | 8.5 | 0.004 |
| F-D | SY | RRW | qRRW12.1 | RM20A | 12 | 3.2 | 12.1 | 0.049 | 7.7 | 0.003 |
| F-D | SY | RRW | qRRW12.4 | RM511 | 12 | 59.8 | 12.4 | 0.007 | 14.1 | -0.004 |
| F-D | MY | RRW | qRRW1.3 | RM576 | 1 | 51 | 1.3 | 0.047 | 8.2 | 0.003 |
| F-D | MY | RRW | qRRW4.4 | RM142 | 4 | 68.5 | 4.4 | 0.016 | 11.5 | -0.003 |
| F-D | SY | RSW | qRSW1.2 | RM283 | 1 | 31.4 | 1.2 | 0.036 | 8.5 | 0.012 |
| F-D | MY | RSW | qRSW3.8 | RM16 | 3 | 131.5 | 3.8 | 0.028 | 9.7 | -0.012 |
| F-D | MY | RSW | qRSW5.7 | RM26 | 5 | 118.8 | 5.7 | 0.039 | 8.7 | 0.002 |
| F-D | MY | RSW | qRSW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.006 | 14.6 | -0.009 |
| F-D | SY | RTW | qRTW1.2 | RM283 | 1 | 31.4 | 1.2 | 0.039 | 8.4 | 0.015 |
| F-D | SY | RTW | qRTW12.4 | RM511 | 12 | 59.8 | 12.4 | 0.034 | 8.9 | -0.017 |
| F-D | MY | RTW | qRTW3.8 | RM16 | 3 | 131.5 | 3.8 | 0.021 | 10.6 | -0.017 |
| F-D | MY | RTW | qRTW3.12 | RM514 | 3 | 216.4 | 3.12 | 0.043 | 8.2 | -0.007 |
| F-D | MY | RTW | qRTW5.7 | RM26 | 5 | 118.8 | 5.7 | 0.032 | 9.4 | 0.001 |
| F-D | MY | RTW | qRTW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.005 | 15.4 | -0.01 |
| F-D | SY | RWRSR | qRWRSR9.3 | RM219 | 9 | 11.7 | 9.3 | 0.044 | 8 | 0.024 |
| F-D | MY | RWRSR | qRWRSR5.2 | RM574 | 5 | 41 | 5.2 | 0.006 | 14.7 | 0.056 |
| F-D | MY | RWRSR | qRWRSR6.3 | RM510 | 6 | 20.8 | 6.3 | 0.012 | 12.6 | 0.043 |
Table 5 Identification of QTLs for measured traits in two populations in Beijing, China.
| Treatment | Population | Trait | QTL | Marker | Chromosome | Position (cM) | Bin | Probability | R2 (%) | Additive |
|---|---|---|---|---|---|---|---|---|---|---|
| E-D | SY | RRL | qRRL9.3 | RM444 | 9 | 3.3 | 9.3 | 0.014 | 11.5 | 0.397 |
| E-D | SY | RRL | qRRL12.4 | RM511 | 12 | 59.8 | 12.4 | 0.009 | 13 | 0.289 |
| E-D | MY | RRL | qRRL6.6 | RM3307 | 6 | 113.4 | 6.6 | 0.007 | 14.4 | -0.403 |
| E-D | MY | RRL | qRRL9.7 | RM108 | 9 | 73.3 | 9.7 | 0.034 | 11.4 | 0.353 |
| E-D | SY | RRW | qRRW4.1 | RM335 | 4 | 21.5 | 4.1 | 0.019 | 10.9 | 0.009 |
| E-D | SY | RRW | qRRW6.3 | RM510 | 6 | 20.8 | 6.3 | 0.035 | 8.8 | -0.007 |
| E-D | SY | RRW | qRRW7.7 | RM47 | 7 | 90 | 7.7 | 0.036 | 8.8 | -0.009 |
| E-D | SY | RRW | qRRW9.7 | RM160 | 9 | 82.4 | 9.7 | 0.03 | 9.9 | 0.006 |
| E-D | MY | RRW | qRRW2.3 | RM145 | 2 | 49.8 | 2.3 | 0.028 | 9.6 | 0.002 |
| E-D | MY | RRW | qRRW3.12 | RM514 | 3 | 216.4 | 3.12 | 0.049 | 7.9 | -0.002 |
| E-D | MY | RRW | qRRW12.2 | RM155 | 12 | 20.9 | 12.2 | 0.016 | 11.5 | 0.003 |
| E-D | SY | RSW | qRSW12.4 | RM511 | 12 | 59.8 | 12.4 | 0.037 | 8.4 | -0.01 |
| E-D | MY | RSW | qRSW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.032 | 9.2 | -0.004 |
| E-D | MY | RSW | qRSW12.2 | RM155 | 12 | 20.9 | 12.2 | 0.04 | 8.5 | 0.007 |
| E-D | SY | RTW | qRTW7.7 | RM47 | 7 | 90 | 7.7 | 0.039 | 8.7 | -0.024 |
| E-D | SY | RTW | qRTW12.4 | RM511 | 12 | 59.8 | 12.4 | 0.024 | 10.2 | -0.014 |
| E-D | MY | RTW | qRTW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.038 | 8.7 | -0.006 |
| E-D | MY | RTW | qRTW10.6 | RM590 | 10 | 117.2 | 10.6 | 0.046 | 8 | -0.01 |
| E-D | MY | RTW | qRTW12.2 | RM155 | 12 | 20.9 | 12.2 | 0.035 | 9 | 0.01 |
| E-D | SY | RWRSR | qRWRSR2.5 | RM3688 | 2 | 88.2 | 2.5 | 0.02 | 10.8 | -0.019 |
| E-D | SY | RWRSR | qRWRSR6.1 | RM589 | 6 | 3.2 | 6.1 | 0.022 | 10.3 | -0.017 |
| E-D | SY | RWRSR | qRWRSR10.5 | RM184 | 10 | 58 | 10.5 | 0.008 | 13.4 | 0.015 |
| E-D | MY | RWRSR | qRWRSR3.12 | RM514 | 3 | 216.4 | 3.12 | 0.019 | 11 | -0.015 |
| E-D | MY | RWRSR | qRWRSR5.8 | RM538 | 5 | 132.7 | 5.8 | 0.046 | 8 | 0.021 |
| E-D | MY | RWRSR | qRWRSR11.4 | RM287 | 11 | 68.6 | 11.4 | 0.048 | 7.9 | 0.012 |
| E-D | MY | RWRSR | qRWRSR12.2 | RM155 | 12 | 20.9 | 12.2 | 0.005 | 15 | 0.007 |
| F-D | SY | RRL | qRRL2.4 | RM521 | 2 | 62.2 | 2.4 | 0.005 | 14.5 | 0.392 |
| F-D | SY | RRL | qRRL5.6 | RM3295 | 5 | 91.2 | 5.6 | 0.003 | 16.6 | -0.489 |
| F-D | MY | RRL | qRRL4.8 | RM349 | 4 | 146.8 | 4.8 | 0.006 | 15.1 | 0.835 |
| F-D | MY | RRL | qRRL7.1 | RM481 | 7 | 3.2 | 7.1 | 0.015 | 12.1 | 0.166 |
| F-D | SY | RRW | qRRW2.4 | RM290 | 2 | 66 | 2.4 | 0.047 | 8.5 | 0.004 |
| F-D | SY | RRW | qRRW12.1 | RM20A | 12 | 3.2 | 12.1 | 0.049 | 7.7 | 0.003 |
| F-D | SY | RRW | qRRW12.4 | RM511 | 12 | 59.8 | 12.4 | 0.007 | 14.1 | -0.004 |
| F-D | MY | RRW | qRRW1.3 | RM576 | 1 | 51 | 1.3 | 0.047 | 8.2 | 0.003 |
| F-D | MY | RRW | qRRW4.4 | RM142 | 4 | 68.5 | 4.4 | 0.016 | 11.5 | -0.003 |
| F-D | SY | RSW | qRSW1.2 | RM283 | 1 | 31.4 | 1.2 | 0.036 | 8.5 | 0.012 |
| F-D | MY | RSW | qRSW3.8 | RM16 | 3 | 131.5 | 3.8 | 0.028 | 9.7 | -0.012 |
| F-D | MY | RSW | qRSW5.7 | RM26 | 5 | 118.8 | 5.7 | 0.039 | 8.7 | 0.002 |
| F-D | MY | RSW | qRSW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.006 | 14.6 | -0.009 |
| F-D | SY | RTW | qRTW1.2 | RM283 | 1 | 31.4 | 1.2 | 0.039 | 8.4 | 0.015 |
| F-D | SY | RTW | qRTW12.4 | RM511 | 12 | 59.8 | 12.4 | 0.034 | 8.9 | -0.017 |
| F-D | MY | RTW | qRTW3.8 | RM16 | 3 | 131.5 | 3.8 | 0.021 | 10.6 | -0.017 |
| F-D | MY | RTW | qRTW3.12 | RM514 | 3 | 216.4 | 3.12 | 0.043 | 8.2 | -0.007 |
| F-D | MY | RTW | qRTW5.7 | RM26 | 5 | 118.8 | 5.7 | 0.032 | 9.4 | 0.001 |
| F-D | MY | RTW | qRTW7.1 | RM481 | 7 | 3.2 | 7.1 | 0.005 | 15.4 | -0.01 |
| F-D | SY | RWRSR | qRWRSR9.3 | RM219 | 9 | 11.7 | 9.3 | 0.044 | 8 | 0.024 |
| F-D | MY | RWRSR | qRWRSR5.2 | RM574 | 5 | 41 | 5.2 | 0.006 | 14.7 | 0.056 |
| F-D | MY | RWRSR | qRWRSR6.3 | RM510 | 6 | 20.8 | 6.3 | 0.012 | 12.6 | 0.043 |
| Type | Population | Material | Environment | HD | PH | PN | PL | FGP | SNP | SF | TGW | GY | BI | HI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (d) | (cm) | (cm) | (%) | (g) | (g) | (g) | ||||||||
| Type 1 | MY | BL151 | Langfang, 2013 | 118.7 | 109.3 | 5.1 | 27.4 | 204.6 | 230.2 | 88.5 | 29 | 18.6 | 43 | 0.43 |
| Hefei, 2012 | 94 | 119.4 | 6.9 | 26.8 | 109.7 | 160.6 | 68.3 | 29.1 | 21.7 | |||||
| MY | BL158 | Langfang, 2013 | 115.7 | 105.8 | 6.2 | 27.5 | 175.7 | 235 | 74.6 | 29.3 | 23.8 | 51.6 | 0.46 | |
| Hefei, 2012 | 93 | 106.4 | 7.7 | 23.6 | 107.6 | 135.4 | 79.7 | 31.6 | 25.8 | |||||
| MY | BL165 | Langfang, 2013 | 119.3 | 100.6 | 6.7 | 25.5 | 170.4 | 234.6 | 73.1 | 20.6 | 19.5 | 46.7 | 0.42 | |
| Hefei, 2012 | 90 | 115.4 | 8.3 | 24.9 | 92.3 | 126.9 | 72.5 | 30.1 | 22.8 | |||||
| MY | BL167 | Langfang, 2013 | 115.3 | 101.2 | 6.3 | 25.2 | 143.6 | 203.8 | 70.5 | 28.5 | 20.9 | 41.1 | 0.51 | |
| Hefei, 2012 | 94 | 107.7 | 8.3 | 24.2 | 105.2 | 140.6 | 74.9 | 25.9 | 22.7 | |||||
| MY | BL170 | Langfang, 2013 | 116.7 | 108.1 | 4.7 | 27.6 | 239 | 272.6 | 87.7 | 26.9 | 18.7 | 37.3 | 0.5 | |
| Hefei, 2012 | 93 | 120.4 | 6 | 23.7 | 97.7 | 125.2 | 78 | 27.6 | 16.2 | |||||
| MY | BL197 | Langfang, 2013 | 117.7 | 93.2 | 4.5 | 27.2 | 170 | 214.8 | 79.1 | 25.3 | 18.5 | 37.1 | 0.5 | |
| Hefei, 2012 | 92 | 108 | 7.7 | 25.9 | 92 | 150.1 | 61.4 | 30.3 | 21.4 | |||||
| MY | BL198 | Langfang, 2013 | 117.3 | 100.4 | 5.8 | 27.4 | 202.8 | 256.5 | 77.9 | 28 | 22.4 | 45.7 | 0.49 | |
| Hefei, 2012 | 92 | 111.9 | 5.3 | 24.3 | 82.5 | 142.3 | 58.9 | 28.5 | 12.5 | |||||
| Type 2 | SY | BL66 | Langfang, 2013 | 116.7 | 105.9 | 6.7 | 24.9 | 180.5 | 221.2 | 81.6 | 29.2 | 29.2 | 60 | 0.49 |
| Hefei, 2012 | 92 | 109.5 | 7.4 | 24.4 | 116.8 | 148.8 | 78.6 | 30.9 | 26.5 | |||||
| MY | BL152 | Langfang, 2013 | 119 | 101.9 | 6.2 | 25.7 | 216.4 | 248.9 | 87 | 25.7 | 26.3 | 49.9 | 0.53 | |
| Hefei, 2012 | 94 | 111.5 | 7.8 | 26.7 | 96.2 | 142.1 | 67.9 | 30 | 22.4 | |||||
| MY | BL162 | Langfang, 2013 | 117.3 | 107.8 | 5.7 | 29 | 211.3 | 245.3 | 86.2 | 27 | 26.2 | 51 | 0.51 | |
| Hefei, 2012 | 94 | 109.7 | 6.4 | 27.7 | 113.6 | 157.8 | 71.9 | 28.5 | 20.4 | |||||
| MY | BL172 | Langfang, 2013 | 116.7 | 111 | 5.7 | 27.2 | 139.5 | 169.2 | 82.5 | 27.8 | 18.2 | 37 | 0.49 | |
| Hefei, 2012 | 91 | 109.6 | 8.7 | 24.7 | 87.2 | 108.9 | 80 | 30 | 22.7 | |||||
| MY | BL190 | Langfang, 2013 | 117.3 | 110.3 | 5.1 | 27.6 | 171.9 | 208.8 | 82.6 | 29.5 | 23 | 46.9 | 0.49 | |
| Hefei, 2012 | 91 | 115.2 | 8.4 | 24.9 | 103.3 | 140.6 | 73.6 | 30.6 | 26.3 | |||||
| MY | BL192 | Langfang, 2013 | 120.3 | 97.2 | 4.5 | 25.5 | 174.3 | 217.4 | 79.7 | 26.3 | 19.5 | 40.3 | 0.48 | |
| Hefei, 2012 | 95 | 102.4 | 8 | 23 | 85.7 | 131.5 | 65.2 | 28.7 | 19.6 | |||||
| MY | BL195 | Langfang, 2013 | 118 | 106.9 | 5.1 | 28.2 | 150.1 | 225.4 | 66.4 | 27.8 | 17.6 | 34.6 | 0.51 | |
| Hefei, 2012 | 93 | 115.7 | 6.2 | 25.2 | 84 | 138.8 | 60.4 | 29.6 | 15.6 | |||||
| Shuhui 527 | Langfang, 2013 | 116.3 | 98.1 | 4.6 | 25.6 | 130.3 | 162.1 | 80.6 | 27.6 | 16.2 | 34.1 | 0.48 | ||
| Hefei, 2012 | 92.4 | 99.3 | 8 | 24.7 | 97.7 | 127.9 | 77.2 | 29.9 | 21.3 | |||||
| Minghui 86 | Langfang, 2013 | 113.7 | 92.7 | 4.3 | 23.4 | 120.8 | 159.5 | 75.8 | 23.8 | 9.1 | 24.3 | 0.37 | ||
| Hefei, 2012 | 93.8 | 106.5 | 8.1 | 26.3 | 82.8 | 143.7 | 61.3 | 27.8 | 16.3 |
Table 6 Trait performance of two IL populations with significant higher yield than the recurrent parents selected under low phosphorus stress conditions in Langfang, Hebei Province and Hefei, Anhui Province, China.
| Type | Population | Material | Environment | HD | PH | PN | PL | FGP | SNP | SF | TGW | GY | BI | HI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (d) | (cm) | (cm) | (%) | (g) | (g) | (g) | ||||||||
| Type 1 | MY | BL151 | Langfang, 2013 | 118.7 | 109.3 | 5.1 | 27.4 | 204.6 | 230.2 | 88.5 | 29 | 18.6 | 43 | 0.43 |
| Hefei, 2012 | 94 | 119.4 | 6.9 | 26.8 | 109.7 | 160.6 | 68.3 | 29.1 | 21.7 | |||||
| MY | BL158 | Langfang, 2013 | 115.7 | 105.8 | 6.2 | 27.5 | 175.7 | 235 | 74.6 | 29.3 | 23.8 | 51.6 | 0.46 | |
| Hefei, 2012 | 93 | 106.4 | 7.7 | 23.6 | 107.6 | 135.4 | 79.7 | 31.6 | 25.8 | |||||
| MY | BL165 | Langfang, 2013 | 119.3 | 100.6 | 6.7 | 25.5 | 170.4 | 234.6 | 73.1 | 20.6 | 19.5 | 46.7 | 0.42 | |
| Hefei, 2012 | 90 | 115.4 | 8.3 | 24.9 | 92.3 | 126.9 | 72.5 | 30.1 | 22.8 | |||||
| MY | BL167 | Langfang, 2013 | 115.3 | 101.2 | 6.3 | 25.2 | 143.6 | 203.8 | 70.5 | 28.5 | 20.9 | 41.1 | 0.51 | |
| Hefei, 2012 | 94 | 107.7 | 8.3 | 24.2 | 105.2 | 140.6 | 74.9 | 25.9 | 22.7 | |||||
| MY | BL170 | Langfang, 2013 | 116.7 | 108.1 | 4.7 | 27.6 | 239 | 272.6 | 87.7 | 26.9 | 18.7 | 37.3 | 0.5 | |
| Hefei, 2012 | 93 | 120.4 | 6 | 23.7 | 97.7 | 125.2 | 78 | 27.6 | 16.2 | |||||
| MY | BL197 | Langfang, 2013 | 117.7 | 93.2 | 4.5 | 27.2 | 170 | 214.8 | 79.1 | 25.3 | 18.5 | 37.1 | 0.5 | |
| Hefei, 2012 | 92 | 108 | 7.7 | 25.9 | 92 | 150.1 | 61.4 | 30.3 | 21.4 | |||||
| MY | BL198 | Langfang, 2013 | 117.3 | 100.4 | 5.8 | 27.4 | 202.8 | 256.5 | 77.9 | 28 | 22.4 | 45.7 | 0.49 | |
| Hefei, 2012 | 92 | 111.9 | 5.3 | 24.3 | 82.5 | 142.3 | 58.9 | 28.5 | 12.5 | |||||
| Type 2 | SY | BL66 | Langfang, 2013 | 116.7 | 105.9 | 6.7 | 24.9 | 180.5 | 221.2 | 81.6 | 29.2 | 29.2 | 60 | 0.49 |
| Hefei, 2012 | 92 | 109.5 | 7.4 | 24.4 | 116.8 | 148.8 | 78.6 | 30.9 | 26.5 | |||||
| MY | BL152 | Langfang, 2013 | 119 | 101.9 | 6.2 | 25.7 | 216.4 | 248.9 | 87 | 25.7 | 26.3 | 49.9 | 0.53 | |
| Hefei, 2012 | 94 | 111.5 | 7.8 | 26.7 | 96.2 | 142.1 | 67.9 | 30 | 22.4 | |||||
| MY | BL162 | Langfang, 2013 | 117.3 | 107.8 | 5.7 | 29 | 211.3 | 245.3 | 86.2 | 27 | 26.2 | 51 | 0.51 | |
| Hefei, 2012 | 94 | 109.7 | 6.4 | 27.7 | 113.6 | 157.8 | 71.9 | 28.5 | 20.4 | |||||
| MY | BL172 | Langfang, 2013 | 116.7 | 111 | 5.7 | 27.2 | 139.5 | 169.2 | 82.5 | 27.8 | 18.2 | 37 | 0.49 | |
| Hefei, 2012 | 91 | 109.6 | 8.7 | 24.7 | 87.2 | 108.9 | 80 | 30 | 22.7 | |||||
| MY | BL190 | Langfang, 2013 | 117.3 | 110.3 | 5.1 | 27.6 | 171.9 | 208.8 | 82.6 | 29.5 | 23 | 46.9 | 0.49 | |
| Hefei, 2012 | 91 | 115.2 | 8.4 | 24.9 | 103.3 | 140.6 | 73.6 | 30.6 | 26.3 | |||||
| MY | BL192 | Langfang, 2013 | 120.3 | 97.2 | 4.5 | 25.5 | 174.3 | 217.4 | 79.7 | 26.3 | 19.5 | 40.3 | 0.48 | |
| Hefei, 2012 | 95 | 102.4 | 8 | 23 | 85.7 | 131.5 | 65.2 | 28.7 | 19.6 | |||||
| MY | BL195 | Langfang, 2013 | 118 | 106.9 | 5.1 | 28.2 | 150.1 | 225.4 | 66.4 | 27.8 | 17.6 | 34.6 | 0.51 | |
| Hefei, 2012 | 93 | 115.7 | 6.2 | 25.2 | 84 | 138.8 | 60.4 | 29.6 | 15.6 | |||||
| Shuhui 527 | Langfang, 2013 | 116.3 | 98.1 | 4.6 | 25.6 | 130.3 | 162.1 | 80.6 | 27.6 | 16.2 | 34.1 | 0.48 | ||
| Hefei, 2012 | 92.4 | 99.3 | 8 | 24.7 | 97.7 | 127.9 | 77.2 | 29.9 | 21.3 | |||||
| Minghui 86 | Langfang, 2013 | 113.7 | 92.7 | 4.3 | 23.4 | 120.8 | 159.5 | 75.8 | 23.8 | 9.1 | 24.3 | 0.37 | ||
| Hefei, 2012 | 93.8 | 106.5 | 8.1 | 26.3 | 82.8 | 143.7 | 61.3 | 27.8 | 16.3 |
| Trait | Marker | Chromosome | Position | Bin | QTL | Treatment | Langfang, Hebei Province | Hefei, Anhui Province | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (cM) | Probability | R2 (%) | Additive | Probability | R2 (%) | Additive | ||||||||||
| TGW | RM335 | 4 | 22 | 4.1 | qTGW4.1 | A | 0.025 | 8.6 | -1.1 | 0.01 | 11 | -1.2 | ||||
| C | 0.006 | 12.3 | -1.1 | 0.002 | 16 | -1.4 | ||||||||||
| TGW | RM5463 | 6 | 124.4 | 6.6 | qTGW6.6 | A | 0.008 | 11.9 | -1 | 0.003 | 17 | -1.2 | ||||
| C | 0.004 | 13.7 | -0.9 | 0.02 | 9 | -0.9 | ||||||||||
| TGW | RM481 | 7 | 3.2 | 7.1 | qTGW7.1 | C | 0.032 | 7.7 | 0.8 | 0.05 | 6 | 0.8 | ||||
| TGW | RM219 | 9 | 11.7 | 9.3 | qTGW9.3 | A | 0.017 | 9.5 | -0.8 | 0.01 | 12 | -0.9 | ||||
| C | 0.013 | 10.1 | -0.7 | 0.002 | 15 | -1 | ||||||||||
| GY | RM254 | 11 | 110 | 11.6 | qGY11.6 | A | 0.018 | 9.5 | -1.1 | 0.01 | 11 | -2.3 | ||||
Table 7 Validation of five QTLs identified from Shuhui 527/Yetuozai population in Langfang (2013) and Hefei (2012) in China.
| Trait | Marker | Chromosome | Position | Bin | QTL | Treatment | Langfang, Hebei Province | Hefei, Anhui Province | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (cM) | Probability | R2 (%) | Additive | Probability | R2 (%) | Additive | ||||||||||
| TGW | RM335 | 4 | 22 | 4.1 | qTGW4.1 | A | 0.025 | 8.6 | -1.1 | 0.01 | 11 | -1.2 | ||||
| C | 0.006 | 12.3 | -1.1 | 0.002 | 16 | -1.4 | ||||||||||
| TGW | RM5463 | 6 | 124.4 | 6.6 | qTGW6.6 | A | 0.008 | 11.9 | -1 | 0.003 | 17 | -1.2 | ||||
| C | 0.004 | 13.7 | -0.9 | 0.02 | 9 | -0.9 | ||||||||||
| TGW | RM481 | 7 | 3.2 | 7.1 | qTGW7.1 | C | 0.032 | 7.7 | 0.8 | 0.05 | 6 | 0.8 | ||||
| TGW | RM219 | 9 | 11.7 | 9.3 | qTGW9.3 | A | 0.017 | 9.5 | -0.8 | 0.01 | 12 | -0.9 | ||||
| C | 0.013 | 10.1 | -0.7 | 0.002 | 15 | -1 | ||||||||||
| GY | RM254 | 11 | 110 | 11.6 | qGY11.6 | A | 0.018 | 9.5 | -1.1 | 0.01 | 11 | -2.3 | ||||
| [1] | Carriger S, Vallee D.2007. More crop per drop.Rice Today, 6(2): 10-13. |
| [2] | Chen M Y, Ali J, Fu B Y, Xu J L, Zhao M F, Jiang Y Z, Zhu L H, Shi Y Y, Yao D N, Gao Y M, Li Z K.2011. Detection of drought-related loci in rice at reproductive stage using selected introgressed lines.Agric Sci China, 10(1): 1-8. |
| [3] | Dobermann A, Fairhurst T.2000. Rice: Nutrient Disorders & Nutrient Management. Los Banos: Potash & Phosphate Institute, Potash & Phosphate Institute of Canada and International Rice Research Institute: 1-191. |
| [4] | Gao F Y, Lu X J, Kang H Q, Sun S X, Liu G C, Reng G J.2006. Screening and identification for rice tolerance to Pi deficency at seedling stage.Acta Agron Sin, 32(8): 1151-1155. (in Chinese with English abstract) |
| [5] | Guo T R, Yao P C, Zhang Z D, Wang J J, Wang M.2013. Involvement of antioxidative defense system in rice seedlings exposed to aluminum toxicity and phosphorus deficiency.Chin J Rice Sci, 27(6): 653-657. (in Chinese with English abstract) |
| [6] | He Y X, Zheng T Q, Hao X B, Wang L F, Gao Y M, Hua Z T, Zhai H Q, Xu J L, Xu Z J, Zhu L H, Li Z K.2010. Yield performances of japonica introgression lines selected for drought tolerance in a BC breeding programme.Plant Breeding, 129(2): 167-175. |
| [7] | Huang L F, Tao X T, Gao W, Wang Y L, Zhuang H Y.2014. Effect of reduced phosphorus fertilizer application on yield formation and quality of japonica rice in Jiangsu coastal region.Chin J Rice Sci, 28(6): 632-638. (in Chinese with English abstract) |
| [8] | Huang X H, Wei X H, Sang T, Zhao Q, Feng Q, Zhao Y, Li C Y, Zhu C R, Lu T T, Zhang Z W, Li M, Fan D L, Guo Y L, Wang A, Wang L, Deng L W, Li W J, Lu Y Q, Weng Q J, Liu K Y, Huang T, Zhou T Y, Jing Y F, Li W, Lin Z, Buckler E S, Qian Q, Zhang Q F, Li J Y, Han B.2010. Genome-wide association studies of 14 agronomic traits in rice landraces.Nat Genet, 42(11): 961-967. |
| [9] | Ismail A M, Heuer S, Thomson M J, Wissuwa M.2007. Genetic and genomic approaches to develop rice germplasm for problem soils.Plant Mol Biol, 65(4): 547-570. |
| [10] | Lea P J, Miflin B J.2011. Nitrogen assimilation and its relevance to crop improvement. In: Foyer C H, Zhang H. Annual Plant Reviews. West Sussex, UK: Blackwell Publishing Ltd: 42: 1-40. |
| [11] | Li Z K.2005. Strategies for molecular rice breeding in China.Mol Plant Breeding, 3(5): 603-608. (in Chinese with English abstract) |
| [12] | Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F.2003. Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis.Plant Physiol, 131(1): 345-358. |
| [13] | Luo L J.2005. Development of near isogenic introgression lines and molecular breeding on rice.Mol Plant Breeding, 3(5): 609-612. (in Chinese with English abstract) |
| [14] | Lynch J P.2011. Root phenes for enhanced soil exploration and phosphorus acquisition: Tools for future crops.Plant Physiol, 156: 1041-1049. |
| [15] | Ma G H, Yuan L P.2015. Hybrid rice achievements, development and prospect in China.Agric Sci China, 14(2): 197-205. |
| [16] | Meng L J, Lin X Y, Wang J M, Chen K, Cui Y R, Xu J L, Li Z K.2013. Simultaneous improvement of cold tolerance and yield of temperate japonica rice (Oryza sativa L.) by introgression breeding.Plant Breeding, 132(6): 604-612. |
| [17] | Moncada P, Martinez C P, Borrero J, Chatel M, Gauch Jr H, Guimaraes E, Tohme J, McCouch S R.2001. Quantitative trait loci for yield and yield components in an Oryza sativa-Oryza rufipogon BC2F2 population evaluated in an upland environment.Theor Appl Genet, 102(1): 41-52. |
| [18] | Mu P, Huang C, Li J X, Liu L F, Liu U J, Li Z C.2008. QTL yield trait variation and QTL mapping in a DH population of rice under phosphorus deficiency.Acta Agron Sin, 34(7): 1137-1142. (in Chinese with English abstract) |
| [19] | SAS Institute Inc.2008. SAS/STAT Software V9.2 User’s Guide. Cary, North Carolina, USA: SAS Institute Inc: 2430-2611. |
| [20] | Tanksley S D, Nelson J C.1996. Advanced backcross QTL analysis: A method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines.Theor Appl Genet, 92(2): 191-203. |
| [21] | Vinod K K, Heuer S.2012. Approaches towards nitrogen- and phosphorus-efficient rice.AoB Plants, doi: 10.1093/aobpla/pls028. |
| [22] | Wu P.2013. The research on the molecular mechanism of absorption and utilization of high phosphorus in plant and its application.Chin Sci Technol Achiev, 9: 20. (in Chinese) |
| [23] | Xu J L, Gao Y M, Fu B Y, Li Z K.2005. Identification and screening of favorable genes from rice germplasm in backcross introgression populations.Mol Plant Breeding, 3(5): 619-628. (in Chinese with English abstract) |
| [24] | Zang J P, Sun Y, Wang Y, Yang J, Li F, Zhou Y L, Zhu L H, Jessica R, Mohammadhosein F, Xu J L, Li Z K.2008. Dissection of genetic overlap of salt tolerance QTL at the seedling and tillering stages using backcross introgression lines in rice.Sci China: Life Sci, 51(7): 583-591. |
| [25] | Zhang F, Hao X B, Gao Y M, Hua Z T, Ma X F, Chen W F, Xu Z J, Zhu L H, Li Z K.2007. Improving seedling cold tolerance of japonica rice by using the ‘hidden diversity’ in indica rice germplasm in a backcross breeding program.Acta Agron Sin, 33(10): 1618-1624. (in Chinese with English abstract) |
| [26] | Zheng T Q, Xu J L, Fu B Y, Gao Y M, Veruka S, Lafitte R, Zhai H Q, Wan J M, Zhu L H, Li Z K.2007. Application of genetic hitch-hiking and ANOVA in identification of loci for drought tolerance in populations of rice from directional selection.Acta Agron Sin, 33(5): 799-804. (in Chinese with English abstract) |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||