
Rice Science ›› 2016, Vol. 23 ›› Issue (4): 211-218.DOI: 10.1016/j.rsci.2016.06.004
收稿日期:2015-06-08
接受日期:2015-12-14
出版日期:2016-07-28
发布日期:2016-04-11
. [J]. Rice Science, 2016, 23(4): 211-218.
| Serial No. | Accession name | Accession No. a | Origin |
|---|---|---|---|
| KS1 | Kartik Sail | 3243 | Sylhet |
| KS2 | Kartik Sail | 776 | Chittagong |
| KS3 | Katih Shail | 438 | Rajshahi |
| KS4 | Kartik Sail | 539 | Rangpur |
| KS5 | Kartik Sail | 77 | Dhaka |
| KS6 | Kati Shail | 170 | Tangail |
| KS7 | Kartik Sail | 3662 | Sherpur |
| KS8 | Kati Shail | 3631 | Rajshahi |
| KS9 | Kartika | 4053 | Sylhet |
| KS10 | Kartik Sail | 4881 | Tangail |
| KS11 | Kartik Sail | 76 | Dhaka |
| KS12 | Kati Shail | 437 | Rajshahi |
| KS13 | Kartik Sail | 78 | Dhaka |
| KS14 | Kartik Sail | 1882 | Kishorganj |
| KS15 | Kartik sail (2) | 689 | Comilla |
| KS16 | Kartik sail (2) | 846 | Sylhet |
| KS17 | Kartik Sail | 664 | Comilla |
| KS18 | Kartik Sail | 1887 | Kishorganj |
| KS19 | Kartik Sail | 844 | Sylhet |
| KS20 | Katih Shail | 994 | Khulna |
| KS21 | Kartik Sail | 845 | Sylhet |
| DB1 | Dhali Boro | 2250 | Sylhet |
| DB2 | Dhali Boro | 2247 | Sylhet |
| DB3 | Dhali Boro | 2249 | Sylhet |
| DB4 | Dholi Boro | 180 | Tangail |
| DB5 | Dhali Boro | 2245 | Sylhet |
| DB6 | Dholi Boro | 4396 | Sylhet |
| DB7 | Dhali Boro | 2246 | Sylhet |
| DB8 | Dhali Boro | 2244 | Sylhet |
| DB9 | Dhali Boro | 2248 | Sylhet |
| DB10 | Dhali Boro | 2243 | Sylhet |
Table 1 List of 31 duplicate and similar named rice germplasms of Bangladesh.
| Serial No. | Accession name | Accession No. a | Origin |
|---|---|---|---|
| KS1 | Kartik Sail | 3243 | Sylhet |
| KS2 | Kartik Sail | 776 | Chittagong |
| KS3 | Katih Shail | 438 | Rajshahi |
| KS4 | Kartik Sail | 539 | Rangpur |
| KS5 | Kartik Sail | 77 | Dhaka |
| KS6 | Kati Shail | 170 | Tangail |
| KS7 | Kartik Sail | 3662 | Sherpur |
| KS8 | Kati Shail | 3631 | Rajshahi |
| KS9 | Kartika | 4053 | Sylhet |
| KS10 | Kartik Sail | 4881 | Tangail |
| KS11 | Kartik Sail | 76 | Dhaka |
| KS12 | Kati Shail | 437 | Rajshahi |
| KS13 | Kartik Sail | 78 | Dhaka |
| KS14 | Kartik Sail | 1882 | Kishorganj |
| KS15 | Kartik sail (2) | 689 | Comilla |
| KS16 | Kartik sail (2) | 846 | Sylhet |
| KS17 | Kartik Sail | 664 | Comilla |
| KS18 | Kartik Sail | 1887 | Kishorganj |
| KS19 | Kartik Sail | 844 | Sylhet |
| KS20 | Katih Shail | 994 | Khulna |
| KS21 | Kartik Sail | 845 | Sylhet |
| DB1 | Dhali Boro | 2250 | Sylhet |
| DB2 | Dhali Boro | 2247 | Sylhet |
| DB3 | Dhali Boro | 2249 | Sylhet |
| DB4 | Dholi Boro | 180 | Tangail |
| DB5 | Dhali Boro | 2245 | Sylhet |
| DB6 | Dholi Boro | 4396 | Sylhet |
| DB7 | Dhali Boro | 2246 | Sylhet |
| DB8 | Dhali Boro | 2244 | Sylhet |
| DB9 | Dhali Boro | 2248 | Sylhet |
| DB10 | Dhali Boro | 2243 | Sylhet |
| Cluster | No. of genotypes | Serial No. |
|---|---|---|
| I | 11 | KS1, KS2, KS5, KS7, KS9, KS11, KS14, KS17, KS18, KS19, KS21 |
| II | 3 | KS6, KS8, KS13 |
| III | 7 | KS3, KS4, KS10, KS12, KS15, KS16, KS20 |
| IV | 10 | DB1, DB2, DB3, DB4, DB5, DB6, DB7, DB8, DB9, DB10 |
Table 2 Distribution of 31 duplicate and similar named rice germplasms into 4 clusters for 18 morpho-physicochemical characters.
| Cluster | No. of genotypes | Serial No. |
|---|---|---|
| I | 11 | KS1, KS2, KS5, KS7, KS9, KS11, KS14, KS17, KS18, KS19, KS21 |
| II | 3 | KS6, KS8, KS13 |
| III | 7 | KS3, KS4, KS10, KS12, KS15, KS16, KS20 |
| IV | 10 | DB1, DB2, DB3, DB4, DB5, DB6, DB7, DB8, DB9, DB10 |
| Cluster | I | II | III | IV |
|---|---|---|---|---|
| I | 0.93 | |||
| II | 6.91 | 0.69 | ||
| III | 5.07 | 10.21 | 0.74 | |
| IV | 24.61 | 22.17 | 22.06 | 0.62 |
Table 3 Average intra- (bold) and inter-cluster distances (D2) for 18 morpho-physicochemical characters of 31 duplicate and similar named rice germplasms.
| Cluster | I | II | III | IV |
|---|---|---|---|---|
| I | 0.93 | |||
| II | 6.91 | 0.69 | ||
| III | 5.07 | 10.21 | 0.74 | |
| IV | 24.61 | 22.17 | 22.06 | 0.62 |
| Character | Cluster I | Cluster II | Cluster III | Cluster IV | Vector I | Vector II | Combined ranking a |
|---|---|---|---|---|---|---|---|
| Seedling height (cm) | 66 | 69.9 | 56.7 | 34.8 | 0.1093 | -0.0729 | 9 |
| Plant height (cm) | 123.9 | 125.6 | 111.1 | 106.1 | -0.1082 | 0.0332 | 12 |
| Culm diameter (mm) | 5.27 | 5.16 | 4.77 | 4.28 | 1.7199 | -1.788 | 11 |
| Panicle exsertion (mm) | 0.64 | 0.53 | 0.44 | 6.13 | -1.6266 | -0.2472 | 18 |
| Effective tillering number per hill | 11.8 | 11 | 12.1 | 10.3 | 0.2065 | 0.2738 | 3 |
| Days to maturity (d) | 143 | 141 | 142 | 160 | -0.2783 | -0.1692 | 14 |
| Primary branch number per panicle | 10.6 | 9 | 9.1 | 7.1 | 0.847 | 0.1551 | 2 |
| Secondary branch number per panicle | 33.6 | 21.7 | 24.9 | 16.9 | 0.1115 | 0.24 | 6 |
| Grain yield per panicle (g) | 3.2 | 2.7 | 2.3 | 1.9 | 0.2634 | -1.1171 | 17 |
| Grain length (mm) | 8.55 | 8.44 | 8.17 | 7.62 | 0.2474 | -0.3449 | 13 |
| Awn length (mm) | 0.6 | 21.5 | 0 | 30.5 | -0.0198 | 0.1587 | 7 |
| 1000-grain weight (g) | 23.19 | 27.18 | 20.82 | 20.69 | -0.0652 | 0.5106 | 4 |
| Grain yield per hill (g) | 22.42 | 21.28 | 22 | 17.28 | -0.3271 | -0.1641 | 15 |
| Biological yield (g) | 40.9 | 37 | 33.1 | 27.7 | 0.1759 | 0.2617 | 5 |
| Milling rate (%) | 68.2 | 65.3 | 69.9 | 68.7 | 0.0361 | -0.633 | 16 |
| Cooking time (min) | 17 | 20 | 16.5 | 19.1 | -0.4281 | 0.3856 | 10 |
| Amylose content (%) | 25.3 | 25.27 | 25.76 | 21.17 | 0.2999 | -0.2623 | 8 |
| Protein content (%) | 7.65 | 9.03 | 7.92 | 9.12 | 0.8121 | 0.6391 | 1 |
Table 4 Means and latent vectors for 18 morpho-physicochemical characters of 31 duplicate and similar named rice germplasms.
| Character | Cluster I | Cluster II | Cluster III | Cluster IV | Vector I | Vector II | Combined ranking a |
|---|---|---|---|---|---|---|---|
| Seedling height (cm) | 66 | 69.9 | 56.7 | 34.8 | 0.1093 | -0.0729 | 9 |
| Plant height (cm) | 123.9 | 125.6 | 111.1 | 106.1 | -0.1082 | 0.0332 | 12 |
| Culm diameter (mm) | 5.27 | 5.16 | 4.77 | 4.28 | 1.7199 | -1.788 | 11 |
| Panicle exsertion (mm) | 0.64 | 0.53 | 0.44 | 6.13 | -1.6266 | -0.2472 | 18 |
| Effective tillering number per hill | 11.8 | 11 | 12.1 | 10.3 | 0.2065 | 0.2738 | 3 |
| Days to maturity (d) | 143 | 141 | 142 | 160 | -0.2783 | -0.1692 | 14 |
| Primary branch number per panicle | 10.6 | 9 | 9.1 | 7.1 | 0.847 | 0.1551 | 2 |
| Secondary branch number per panicle | 33.6 | 21.7 | 24.9 | 16.9 | 0.1115 | 0.24 | 6 |
| Grain yield per panicle (g) | 3.2 | 2.7 | 2.3 | 1.9 | 0.2634 | -1.1171 | 17 |
| Grain length (mm) | 8.55 | 8.44 | 8.17 | 7.62 | 0.2474 | -0.3449 | 13 |
| Awn length (mm) | 0.6 | 21.5 | 0 | 30.5 | -0.0198 | 0.1587 | 7 |
| 1000-grain weight (g) | 23.19 | 27.18 | 20.82 | 20.69 | -0.0652 | 0.5106 | 4 |
| Grain yield per hill (g) | 22.42 | 21.28 | 22 | 17.28 | -0.3271 | -0.1641 | 15 |
| Biological yield (g) | 40.9 | 37 | 33.1 | 27.7 | 0.1759 | 0.2617 | 5 |
| Milling rate (%) | 68.2 | 65.3 | 69.9 | 68.7 | 0.0361 | -0.633 | 16 |
| Cooking time (min) | 17 | 20 | 16.5 | 19.1 | -0.4281 | 0.3856 | 10 |
| Amylose content (%) | 25.3 | 25.27 | 25.76 | 21.17 | 0.2999 | -0.2623 | 8 |
| Protein content (%) | 7.65 | 9.03 | 7.92 | 9.12 | 0.8121 | 0.6391 | 1 |
| Chromosome | Marker | Position | No. of alleles | Size | Major allele | No. of | No. of | Gene | Polymorphism information content | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (cM) | (bp) | Size (bp) | Frequency (%) | rare alleles | null alleles | diversity | |||||
| 1 | RM283 | 31.4 | 5 | 141-150 | 150 | 44 | 4 | 1 | 0.62 | 0.55 | |
| RM259 | 54.2 | 6 | 166-187 | 170 | 39 | 4 | 0 | 0.76 | 0.73 | ||
| RM237 | 115.2 | 6 | 116-133 | 130 | 33 | 3 | 1 | 0.78 | 0.75 | ||
| RM302 | 147.8 | 9 | 107-214 | 178 | 31 | 7 | 2 | 0.82 | 0.8 | ||
| RM154 | 4.8 | 8 | 157-183 | 163/167 | 22 | 6 | 0 | 0.84 | 0.82 | ||
| 2 | RM279 | 17.3 | 8 | 135-173 | 166 | 33 | 6 | 3 | 0.82 | 0.8 | |
| RM324 | 66 | 9 | 132-166 | 166 | 28 | 7 | 0 | 0.82 | 0.8 | ||
| RM250 | 170.1 | 5 | 149-155 | 152 | 61 | 4 | 0 | 0.59 | 0.56 | ||
| RM60 | 0 | 10 | 147-182 | 162 | 25 | 8 | 1 | 0.86 | 0.85 | ||
| 3 | RM218 | 67.8 | 10 | 126-152 | 136 | 22 | 8 | 3 | 0.86 | 0.84 | |
| RM55 | 168.2 | 10 | 215-240 | 229 | 17 | 9 | 3 | 0.89 | 0.88 | ||
| RM227 | 214.7 | 4 | 97-105 | 97 | 47 | 1 | 1 | 0.66 | 0.6 | ||
| RM307 | 0 | 6 | 123-177 | 131 | 69 | 5 | 3 | 0.5 | 0.48 | ||
| 4 | RM273 | 94.4 | 7 | 200-217 | 202 | 39 | 4 | 0 | 0.77 | 0.74 | |
| RM241 | 106.2 | 7 | 100-130 | 126 | 44 | 5 | 0 | 0.72 | 0.69 | ||
| RM127 | 150.1 | 4 | 212-224 | 218 | 56 | 1 | 0 | 0.61 | 0.56 | ||
| RM413 | 26.7 | 9 | 68-105 | 68 | 22 | 7 | 2 | 0.86 | 0.85 | ||
| 5 | RM267 | 28.6 | 8 | 150-185 | 166 | 36 | 6 | 0 | 0.75 | 0.71 | |
| RM161 | 96.9 | 6 | 166-182 | 171 | 36 | 3 | 0 | 0.77 | 0.73 | ||
| RM133 | 0 | 4 | 225-232 | 227 | 69 | 2 | 0 | 0.48 | 0.44 | ||
| 6 | RM584 | 26.2 | 10 | 160-214 | 163 | 33 | 8 | 0 | 0.83 | 0.81 | |
| RM541 | 75.5 | 11 | 142-194 | 160 | 25 | 9 | 0 | 0.84 | 0.83 | ||
| RM162 | 108.3 | 6 | 223-233 | 223 | 33 | 3 | 0 | 0.77 | 0.74 | ||
| RM125 | 24.8 | 4 | 130-145 | 141 | 61 | 1 | 0 | 0.56 | 0.51 | ||
| 7 | RM214 | 34.7 | 8 | 110-137 | 117 | 33 | 5 | 0 | 0.78 | 0.75 | |
| RM18 | 90.4 | 8 | 160-180 | 169 | 22 | 5 | 1 | 0.85 | 0.84 | ||
| RM337 | 1.1 | 6 | 162-202 | 168 | 42 | 3 | 0 | 0.71 | 0.67 | ||
| 8 | RM223 | 80.5 | 8 | 138-163 | 155 | 31 | 5 | 0 | 0.82 | 0.8 | |
| RM256 | 101.5 | 8 | 103-138 | 108 | 36 | 5 | 0 | 0.78 | 0.76 | ||
| RM433 | 116 | 13 | 207-248 | 237 | 28 | 12 | 1 | 0.87 | 0.86 | ||
| RM296 | 0 | 8 | 121-139 | 125 | 39 | 6 | 0 | 0.72 | 0.68 | ||
| 9 | RM242 | 73.3 | 8 | 190-214 | 203 | 28 | 6 | 0 | 0.83 | 0.81 | |
| RM215 | 99.4 | 7 | 159-176 | 163 | 31 | 6 | 1 | 0.83 | 0.81 | ||
| RM311 | 25.2 | 10 | 170-201 | 176/182 | 19 | 7 | 0 | 0.86 | 0.84 | ||
| 10 | RM271 | 59.4 | 8 | 81-98 | 93 | 28 | 6 | 1 | 0.82 | 0.8 | |
| RM258 | 70.8 | 13 | 139-167 | 146 | 22 | 11 | 1 | 0.88 | 0.87 | ||
| RM171 | 92.8 | 6 | 314-340 | 314 | 36 | 4 | 0 | 0.75 | 0.72 | ||
| RM21 | 85.7 | 14 | 129-173 | 166 | 22 | 12 | 0 | 0.89 | 0.88 | ||
| 11 | RM229 | 77.8 | 8 | 105-129 | 112 | 25 | 5 | 0 | 0.82 | 0.8 | |
| RM206 | 102.9 | 13 | 116-171 | 157 | 17 | 12 | 0 | 0.9 | 0.89 | ||
| RM224 | 120.1 | 9 | 123-155 | 147 | 36 | 7 | 0 | 0.8 | 0.78 | ||
| RM286 | 0 | 6 | 96-121 | 96 | 31 | 4 | 0 | 0.79 | 0.75 | ||
| 12 | RM19 | 20.9 | 8 | 223-239 | 228 | 28 | 5 | 1 | 0.83 | 0.81 | |
| RM247 | 32.3 | 6 | 126-144 | 136 | 31 | 3 | 0 | 0.78 | 0.75 | ||
| RM277 | 57.2 | 3 | 116-123 | 116/119 | 47 | 1 | 0 | 0.55 | 0.45 | ||
| Minimum | 3 | 68 | 17 | 1 | 0 | 0.48 | 0.44 | ||||
| Average | 7.8 | 161.2 | 35 | 5.8 | 0.6 | 0.77 | 0.74 | ||||
| Maximum | 14 | 314 | 69 | 12 | 3 | 0.9 | 0.89 | ||||
Table 5 Allele variation, gene diversity and polymorphism information content of 45 simple sequence repeat markers across 31 duplicate and similar named rice germplasms.
| Chromosome | Marker | Position | No. of alleles | Size | Major allele | No. of | No. of | Gene | Polymorphism information content | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (cM) | (bp) | Size (bp) | Frequency (%) | rare alleles | null alleles | diversity | |||||
| 1 | RM283 | 31.4 | 5 | 141-150 | 150 | 44 | 4 | 1 | 0.62 | 0.55 | |
| RM259 | 54.2 | 6 | 166-187 | 170 | 39 | 4 | 0 | 0.76 | 0.73 | ||
| RM237 | 115.2 | 6 | 116-133 | 130 | 33 | 3 | 1 | 0.78 | 0.75 | ||
| RM302 | 147.8 | 9 | 107-214 | 178 | 31 | 7 | 2 | 0.82 | 0.8 | ||
| RM154 | 4.8 | 8 | 157-183 | 163/167 | 22 | 6 | 0 | 0.84 | 0.82 | ||
| 2 | RM279 | 17.3 | 8 | 135-173 | 166 | 33 | 6 | 3 | 0.82 | 0.8 | |
| RM324 | 66 | 9 | 132-166 | 166 | 28 | 7 | 0 | 0.82 | 0.8 | ||
| RM250 | 170.1 | 5 | 149-155 | 152 | 61 | 4 | 0 | 0.59 | 0.56 | ||
| RM60 | 0 | 10 | 147-182 | 162 | 25 | 8 | 1 | 0.86 | 0.85 | ||
| 3 | RM218 | 67.8 | 10 | 126-152 | 136 | 22 | 8 | 3 | 0.86 | 0.84 | |
| RM55 | 168.2 | 10 | 215-240 | 229 | 17 | 9 | 3 | 0.89 | 0.88 | ||
| RM227 | 214.7 | 4 | 97-105 | 97 | 47 | 1 | 1 | 0.66 | 0.6 | ||
| RM307 | 0 | 6 | 123-177 | 131 | 69 | 5 | 3 | 0.5 | 0.48 | ||
| 4 | RM273 | 94.4 | 7 | 200-217 | 202 | 39 | 4 | 0 | 0.77 | 0.74 | |
| RM241 | 106.2 | 7 | 100-130 | 126 | 44 | 5 | 0 | 0.72 | 0.69 | ||
| RM127 | 150.1 | 4 | 212-224 | 218 | 56 | 1 | 0 | 0.61 | 0.56 | ||
| RM413 | 26.7 | 9 | 68-105 | 68 | 22 | 7 | 2 | 0.86 | 0.85 | ||
| 5 | RM267 | 28.6 | 8 | 150-185 | 166 | 36 | 6 | 0 | 0.75 | 0.71 | |
| RM161 | 96.9 | 6 | 166-182 | 171 | 36 | 3 | 0 | 0.77 | 0.73 | ||
| RM133 | 0 | 4 | 225-232 | 227 | 69 | 2 | 0 | 0.48 | 0.44 | ||
| 6 | RM584 | 26.2 | 10 | 160-214 | 163 | 33 | 8 | 0 | 0.83 | 0.81 | |
| RM541 | 75.5 | 11 | 142-194 | 160 | 25 | 9 | 0 | 0.84 | 0.83 | ||
| RM162 | 108.3 | 6 | 223-233 | 223 | 33 | 3 | 0 | 0.77 | 0.74 | ||
| RM125 | 24.8 | 4 | 130-145 | 141 | 61 | 1 | 0 | 0.56 | 0.51 | ||
| 7 | RM214 | 34.7 | 8 | 110-137 | 117 | 33 | 5 | 0 | 0.78 | 0.75 | |
| RM18 | 90.4 | 8 | 160-180 | 169 | 22 | 5 | 1 | 0.85 | 0.84 | ||
| RM337 | 1.1 | 6 | 162-202 | 168 | 42 | 3 | 0 | 0.71 | 0.67 | ||
| 8 | RM223 | 80.5 | 8 | 138-163 | 155 | 31 | 5 | 0 | 0.82 | 0.8 | |
| RM256 | 101.5 | 8 | 103-138 | 108 | 36 | 5 | 0 | 0.78 | 0.76 | ||
| RM433 | 116 | 13 | 207-248 | 237 | 28 | 12 | 1 | 0.87 | 0.86 | ||
| RM296 | 0 | 8 | 121-139 | 125 | 39 | 6 | 0 | 0.72 | 0.68 | ||
| 9 | RM242 | 73.3 | 8 | 190-214 | 203 | 28 | 6 | 0 | 0.83 | 0.81 | |
| RM215 | 99.4 | 7 | 159-176 | 163 | 31 | 6 | 1 | 0.83 | 0.81 | ||
| RM311 | 25.2 | 10 | 170-201 | 176/182 | 19 | 7 | 0 | 0.86 | 0.84 | ||
| 10 | RM271 | 59.4 | 8 | 81-98 | 93 | 28 | 6 | 1 | 0.82 | 0.8 | |
| RM258 | 70.8 | 13 | 139-167 | 146 | 22 | 11 | 1 | 0.88 | 0.87 | ||
| RM171 | 92.8 | 6 | 314-340 | 314 | 36 | 4 | 0 | 0.75 | 0.72 | ||
| RM21 | 85.7 | 14 | 129-173 | 166 | 22 | 12 | 0 | 0.89 | 0.88 | ||
| 11 | RM229 | 77.8 | 8 | 105-129 | 112 | 25 | 5 | 0 | 0.82 | 0.8 | |
| RM206 | 102.9 | 13 | 116-171 | 157 | 17 | 12 | 0 | 0.9 | 0.89 | ||
| RM224 | 120.1 | 9 | 123-155 | 147 | 36 | 7 | 0 | 0.8 | 0.78 | ||
| RM286 | 0 | 6 | 96-121 | 96 | 31 | 4 | 0 | 0.79 | 0.75 | ||
| 12 | RM19 | 20.9 | 8 | 223-239 | 228 | 28 | 5 | 1 | 0.83 | 0.81 | |
| RM247 | 32.3 | 6 | 126-144 | 136 | 31 | 3 | 0 | 0.78 | 0.75 | ||
| RM277 | 57.2 | 3 | 116-123 | 116/119 | 47 | 1 | 0 | 0.55 | 0.45 | ||
| Minimum | 3 | 68 | 17 | 1 | 0 | 0.48 | 0.44 | ||||
| Average | 7.8 | 161.2 | 35 | 5.8 | 0.6 | 0.77 | 0.74 | ||||
| Maximum | 14 | 314 | 69 | 12 | 3 | 0.9 | 0.89 | ||||
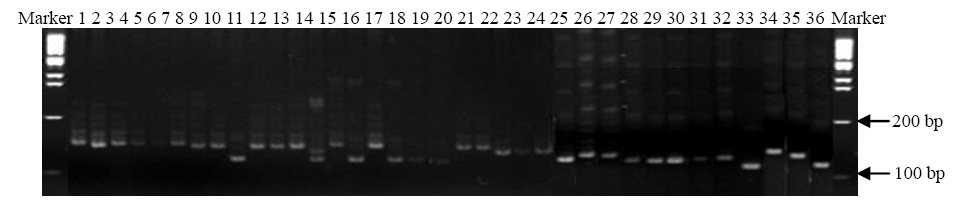
Fig. 2. DNA profile of 36 rice genotypes using RM224 marker.1, KS1; 2, KS2; 3, KS3; 4, KS4; 5, KS5; 6, KS6; 7, KS7; 8, KS8; 9, KS9; 10, KS10; 11, KS11; 12, KS12; 13, KS13; 14, KS14; 15, KS15; 16, KS16; 17, KS17; 18, KS18; 19, KS19; 20, KS20; 21, KS21; 22, BR4; 23, BR23; 24, DB1; 25, DB2; 26, DB3; 27, DB4; 28, DB5; 29, DB6; 30, DB7; 31, DB8; 32, DB9; 33, DB10; 34, BRRI dhan 28; 35, BRRI dhan 29; 36, BR14.
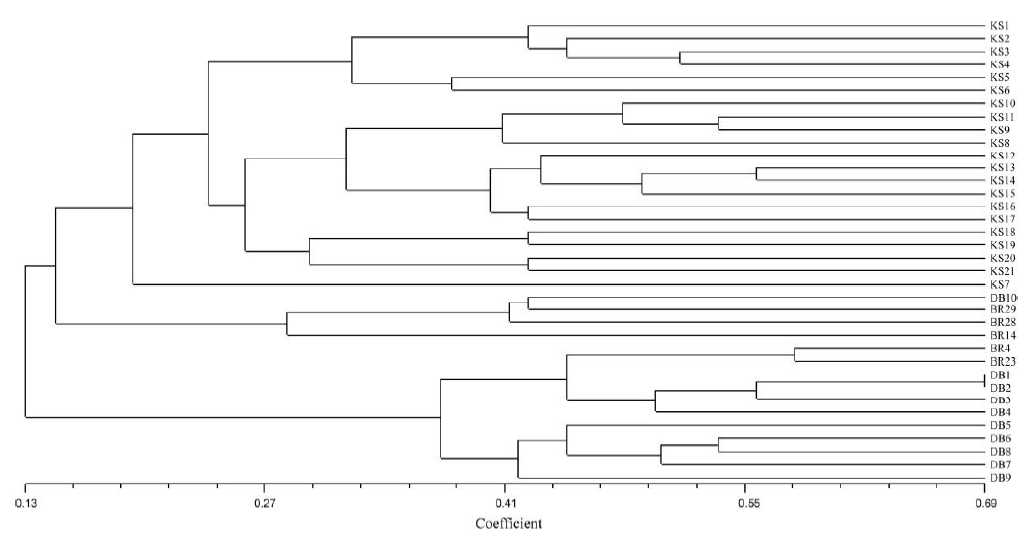
Fig. 3. Dendrogram of 31 duplicate and similar named rice germplasms derived from unweighted pair-group method with arithmetic means cluster analysis based on Nei’s genetic distance across 45 simple sequence repeat markers.BR28, BRRI dhan 28; BR29, BRRI dhan 29.
| 1 | Adair C R.1952. The Mcgill miller method for determining the milling quality of small samples of rice.Rice J, 55(2): 21-23. |
| 2 | Ahmed M S, Akter K, Khalequzzaman M, Rashid E S M H, Bashar M K.2010. Diversity analysis in Boro rice (Oryza sativa L.) accessions.Bangl J Agric Res, 35(1): 29-36. |
| 3 | AOAC.1995. Association of Official Agricultural Chemists. Methods of Analysis. Washington DC, USA. |
| 4 | Chakravorty A, Ghosh P D, Sahu P K.2013. Multivariate analysis of phenotypic diversity of land races of rice of West Bengal.Am J Exp Agric, 3(1): 110-123. |
| 5 | Chen X, Cho Y G, McCouch S R.2002. Sequence divergence of rice microsatellites in Oryza and other plant species.Mol Genet Genom, 268: 331-343. |
| 6 | Collard B C Y, Das A, Virk P S, Mackill D J.2007. Evaluation of quick and dirty DNA extraction methods for marker-assisted selection in rice (Oryza sativa L.).Plant Breeding, 126(1): 47-50. |
| 7 | Datt S, Mani S C.2003. Genetic divergence in elite genotypes of Basmati rice (Oryza sativa L.).Ind J Genet Plant Breeding, 63(1): 73-74. |
| 8 | Fukuoka S, Suu T D, Ebanna K, Trinh L N, Nagamine T, Okuno K.2006. Diversity in phenotypic profiles in land races populations of Vietnamese rice: A case study of agronomic characters for conserving crop genetic diversity on farm.Genet Resour Crop Evol, 53: 753-761. |
| 9 | Hamid A, Nasiruddin M, Haque M, Haque E.1982. Collection of names of the local varieties. In: Taluckdar M H R. Local rice varieties. Joydevpur, Gazipur, Bangladesh: Bangladesh Rice Research Institute. (in Bengali) |
| 10 | Hossain M Z.2008. Genetic diversity study in fine grain and aromatic land races of rice (Oryza sativa L.) by morpho-physico-chemical characters and microsatellite DNA markers. Gazipur, Bangladesh: Bangabandhu Sheikh Mujibur Rahman Agricultual University. |
| 11 | Hossain M, Jaim W M H, Alam M S, Rahman A N M M.2013. Rice biodiversity in Bangladesh: Adoption, diffusion and disappearance of varieties. Dhaka, Bangladesh: BRAC Research and Evalution Division. |
| 12 | Irish B M, Goenaga R, Zhang D, Schnell R J, Brown J S, Motamayor J C.2010. Microsatellite fingerprinting of the USDA-ARS tropical agriculture research station cacao (Theobroma cacao L.) germplasm collection.Crop Sci, 50: 656-667. |
| 13 | Rholf F.2002. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System Version 2.2. New York, USA: Department of Ecology and Evolution, State University of New York. |
| 14 | Joshi R K, Behera L.2006. Identification and differentiation of indigenous non-Basmati aromatic rice genotypes of India using microsatellite markers.Afr J Biotechnol, 6(4): 348-354. |
| 15 | Juilano B O, Nazareno M B, Ramos N.1969. Properties of waxy and isogenic nonwaxy rices differing in starch gelatinization temperature.J Agric Food Chem, 17: 1364-1369. |
| 16 | Juilano B O.1971. A simplified assay for milled rice amylose.Cereal Sci Today, 16: 334-360. |
| 17 | Junjian N I, Colowit P M, Mackill D J.2002. Evaluation of genetic diversity in rice subspecies using microsatellite markers.Crop Sci, 42: 601-607. |
| 18 | Kaushik A, Jain S, McCouch S R, Jain R K.2011. Phylogenetic relationships among various groups of rice (Oryza sativa L.) as revealed by microsatellite and transposable element-based marker analysis.Ind J Genet Plant Breeding, 71(2): 139-150. |
| 19 | Liu K, Muse S V.2005. Power Marker: Integrated analysis environment for genetic marker data.Bioinformatics, 21: 2128-2129. |
| 20 | Lund B, Ortiz R, Skovgaard I M, Waugh R, Andersen S B.2003. Analysis of potential duplicates in barley genebank collections using re-sampling of microsatellite data.Theor Appl Genet, 106(6): 1129-1138. |
| 21 | Masuduzzaman M.2010. Morphological, molecular characterization and Agrobactrium-mediated genetic transformation in jute (Corchorus spp). Mymensingh, Bangladesh: Bangladesh Agricultural University: 41-46. |
| 22 | Nandakumar N, Singh A K, Sharma R K, Mohapatra T, Prabhu K V, Zaman F U.2004. Molecular fingerprinting of hybrids and assessment of genetic purity of hybrid seeds in rice using microsatellite markers.Euphytica, 136(3): 257-264. |
| 23 | Nascimento W F, Silva E F, Veasey E A.2011. Agro- morphological characterization of upland rice accessions.Sci Agric, 68(6): 652-660. |
| 24 | Nei M, Takezaki N.1994. Estimation of genetic distances and phylogenetic trees from DNA analysis. Proceedings, 5th World Congress on Genetics Applied to Livestock Production, University of Guelph, Guelph, Ontario, Canada, 7-12 August. |
| 25 | Rao C R.1952. Advance Statistical Methods in Biometrical Research. New York, USA: John Willey and Sons Inc: 390. |
| 26 | Rao C R.1964. The use and interception of principal component analysis in applied research.Sankhya Ser A, 22: 317-318. |
| 27 | Roy S K, Kundu A, Chand S P, Senapati B K.2004. Diversity of panicle characters in Aman rice (Oryza sativa L.).Environ Ecol, 22: 500-503. |
| 28 | Sohrabi M, Rafii M Y, Hanafi M M, Akmar A S N, Latif M A. 2012. Genetic diversity of upland rice germplasm in Malaysia based on quantitative traits.Sci World J, 2012: 1-9. |
| 29 | Song Z P, Xu X, Wang B, Chen J K, Lu B R.2003. Genetic diversity in the northernmost Oryza rufipogon populations estimated by SSR markers.Theor Appl Genet, 107(8): 1492-1499. |
| 30 | Steel R G D, Torrie J H.1980. Principles and Procedures of Statistics. A Biometrical Approach. 2nd edn. New York, USA: Mcgraw-Hill Book Company Inc: 550. |
| 31 | Thomson M J, Septiningsih E M, Suwardjo F, Santoso T J, Silitonga T S, McCouch S R.2007. Genetic diversity analysis of traditional and improved Indonesian rice (Oryza sativa L.) germplasm using microsatellite markers.Theor Appl Genet, 114(3): 559-568. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||