
Rice Science ›› 2018, Vol. 25 ›› Issue (3): 132-141.DOI: 10.1016/j.rsci.2018.02.004
收稿日期:2017-12-20
接受日期:2018-02-26
出版日期:2018-05-04
发布日期:2018-03-07
. [J]. Rice Science, 2018, 25(3): 132-141.
| Trait | Wild type | sg3 mutant |
|---|---|---|
| GL (mm) (n = 30) | 8.28 ± 0.12 | 10.13 ± 0.10** |
| GW (mm) (n = 30) | 3.17 ± 0.12 | 2.60 ± 0.14** |
| L/W (n = 30) | 2.61 ± 0.10 | 3.89 ± 0.17** |
| GT (mm) (n = 30) | 2.04 ± 0.11 | 1.99 ± 0.14 |
| PL (cm) (n = 10) | 23.16 ± 0.57 | 23.50 ± 0.50 |
| PH (cm) (n = 10) | 93.37 ± 1.80 | 83.12 ± 1.52** |
| PN (n = 10) | 7.33 ± 0.81 | 10.83 ± 0.74** |
| TGW (g) (n = 6) | 26.07 ± 0.12 | 25.92 ± 0.16 |
| NGP (n = 10) | 225.33 ± 10.02 | 216.33 ± 12.85 |
| SSR (%) (n = 10) | 87.02 ± 3.60 | 77.33 ± 4.70** |
| YPP (g) (n = 5) | 32.18 ± 1.31 | 36.90 ± 0.93** |
Table 1 Comparison of agronomic traits between sg3 and wild type.
| Trait | Wild type | sg3 mutant |
|---|---|---|
| GL (mm) (n = 30) | 8.28 ± 0.12 | 10.13 ± 0.10** |
| GW (mm) (n = 30) | 3.17 ± 0.12 | 2.60 ± 0.14** |
| L/W (n = 30) | 2.61 ± 0.10 | 3.89 ± 0.17** |
| GT (mm) (n = 30) | 2.04 ± 0.11 | 1.99 ± 0.14 |
| PL (cm) (n = 10) | 23.16 ± 0.57 | 23.50 ± 0.50 |
| PH (cm) (n = 10) | 93.37 ± 1.80 | 83.12 ± 1.52** |
| PN (n = 10) | 7.33 ± 0.81 | 10.83 ± 0.74** |
| TGW (g) (n = 6) | 26.07 ± 0.12 | 25.92 ± 0.16 |
| NGP (n = 10) | 225.33 ± 10.02 | 216.33 ± 12.85 |
| SSR (%) (n = 10) | 87.02 ± 3.60 | 77.33 ± 4.70** |
| YPP (g) (n = 5) | 32.18 ± 1.31 | 36.90 ± 0.93** |
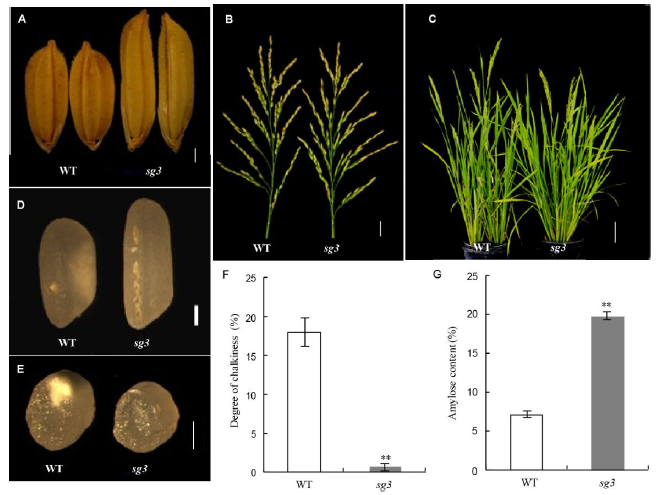
Fig. 1. Comparisons of different traits between wild type (WT) and sg3 mutant. A, Mature grains. Scale bar, 1 mm. B, Mature panicles. Scale bar, 2 cm. C, Plants during anthesis. Scale bar, 10 cm. D, Chalkiness of the polished rice. Scale bar, 1 mm. E, Cross section of the polished rice. Scale bar, 1 mm. F, Degree of chalkiness. G, Amylose content. All data represent Means ± SD (n = 3). ** represents significant difference at the 0.01 level by the Student’s t-test.
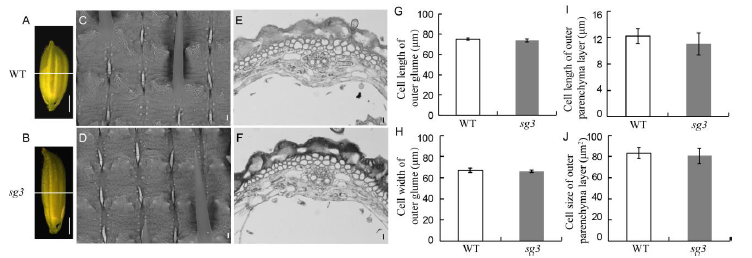
Fig. 2. Spikelet hulls of wild type (WT) and sg3 mutant. A and B, Spikelets of WT and sg3 mutant, respectively. Scale bar, 2 mm. C and D, Glume outer surfaces of WT and sg3 mutant, respectively. Scale bar, 10 μm. E and F, Cross section of outer parenchyma layer from the dotted line position in A and B, respectively. Scale bar, 10 μm. G, Cell length of outer glumes. H, Cell width of outer glumes. I, Cell length of outer parenchyma layer of spikelet hulls. J, Cell size of outer parenchyma layer of spikelet hulls. All data represent Means ± SD (n = 10).
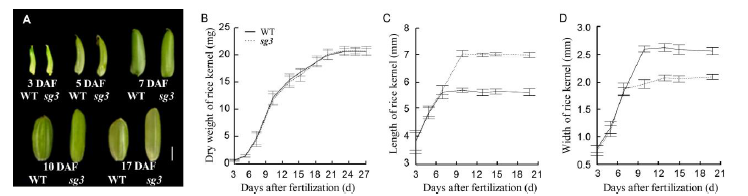
Fig. 3. Development process of rice grains of sg3 mutant and wild type (WT). A, Representative rice grains at 3 days after fertilization (DAF), 5 DAF, 7 DAF, 10 DAF and 17 DAF, respectively. Scale bar, 2 mm. B, Dry weight of rice kernel. C, Length of rice kernel. D, Width of rice kernel. All data represent Means ± SD (n = 20).
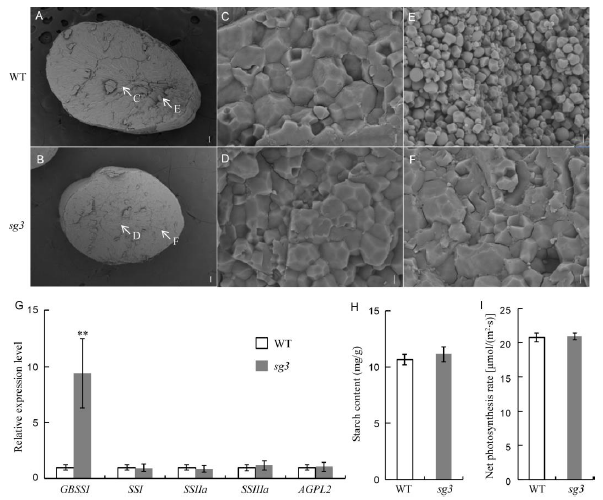
Fig. 4. Scanning electron microscope of the cross section of grains and the physiological changes of wild type (WT) and sg3 mutant. A and B, Cross section of grains. Scale bar, 100 μm. The arrows in A and B show the position of images for C to F. C and D, Starch granules in the central endosperm cell. Scale bar, 3 μm. E and F, Starch granules in the abdominal endosperm cell, Scale bar, 3 μm. G, Relative expression levels of genes related to starch biosynthesis. H, Starch content in the leaf sheath before heading. I, Net photosynthesis rate of flag leaves after heading. All data represent Means ± SD (n = 6). ** represents significant difference at the 0.01 level by the Student’s t-test.
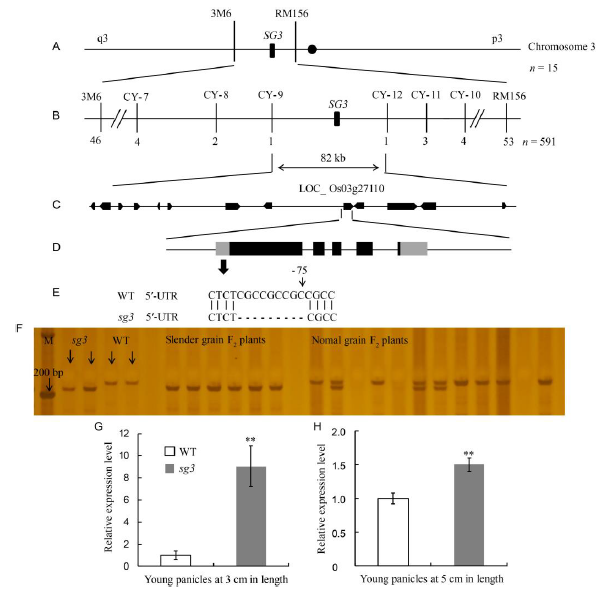
Fig. 5. Map-based cloning of SG3. A, Primary mapping of SG3 on chromosome 3 based on 15 individuals; B, SG3 was fine-mapped to an interval of 82 kb by 591 individuals; C, Thirteen genes were annotated on the 82 kb region; D, Structure of LOC_ Os03g27110; E, Mutation locus in LOC_ Os03g27110; F, The differences in 5′-UTR region of sg3, wild type (WT) and the F2 population derived from sg3/Zhenongda 104; G, LOC_ Os03g27110 expression levels of young panicles at 3 cm in length; H, LOC_ Os03g27110 expression levels of young panicles at 5 cm in length.M, Marker. All data represent Means ± SD (n = 3). ** represents significant difference at the 0.01 level by the Student’s t-test.
| [1] | Abe Y, Mieda K, Ando T, Kono I, Yano M, Kitano H, Iwasaki Y.2010. The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes Genet Syst, 85(5): 327-339. |
| [2] | Bian J M, He H H, Li C J, Shi H, Zhu C L, Peng X S, Fu J R, He X P, Chen X R, Hu L F, Ouyang L J.2013. Identification and validation of a new grain weight QTL in rice.Genet Mol Res, 12(4): 5623-5633. |
| [3] | Calingacion M, Laborte A, Nelson A, Resurreccion A, Concepcion J C, Daygon V D, Mumm R, Reinke R, Dipti S, Bassinello P Z, Manful J, Sophany S, Lara K C, Bao J S, Xie L H, Loaiza K, El-hissewy A, Gayin J, Sharm, N, Rajeswari S, Manonmani S, Rani N S, Kota S, Indrasari S D, Habibi F, Hosseini M, Tavasoli F, Suzuki K, Umemoto T, Boualaphanh C, Lee H H, Hung Y P, Ramli A, Aung P P, Ahmad R, Wattoo J I, Bandonill E, Romero M, Brites C M, Hafeel R, Lur H S, Cheaupun K, Jongdee S, Blanco P, Bryant R, Lang N T, Hall R D, Fitzgerald M.2014. Diversity of global rice markets and the science required for consumer-targeted rice breeding.PLoS One, 9(1): e85106. |
| [4] | Fan C C, Xing Y Z, Mao H L, Lu T T, Han B, Xu C G, Li X H, Zhang Q F.2006. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein.Theor Appl Genet, 112(6): 1164-1171. |
| [5] | Floris M, Mahgoub H, Lanet E, Robaglia C, Menand B.2009. Post-transcriptional regulation of gene expression in plants during abiotic stress.Int J Mol Sci, 10(7): 3168-3185. |
| [6] | Gao Z Y, Zhao S C, He W M, Guo L B, Peng Y L, Wang J J, Guo X S, Zhang X M, Rao Y C, Zhang C, Dong G J, Zheng F Y, Lu C X, Hu J, Zhou Q, Liu H J, Wu H Y, Xu J, Ni P X, Zeng D L, Liu D H, Tian P, Gong L H, Ye C, Zhang G H, Wang J, Tian F K, Xue D W, Liao Y, Zhu L, Chen M S, Li J Y, Cheng S H, Zhang G Y, Wang J, Qian Q.2013. Dissecting yield-associated loci in super hybrid rice by resequencing recombinant inbred lines and improving parental genome sequences.Prol Natl Acad Sci USA, 110(35): 14492-14497. |
| [7] | Haig D.2013. Kin conflict in seed development: An interdependent but fractious collective.Annu Rev Cell Dev Biol, 29(1): 189-211. |
| [8] | Hu J, Wang Y X, Fang Y X, Zeng L J, Xu J, Yu H P, Shi Z Y, Pan J J, Zhang D, Kang S J, Zhu L, Dong G J, Guo L B, Zeng D L, Zhang G H, Xie L H, Xiong G S, Li J Y, Qian Q.2015. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant, 8(10): 1455-1465. |
| [9] | Huang H X, Qian Q.2017. Progress in genetic research of rice grain shape and breeding achievements of long-grain shape and good quality japonica rice. Chin J Rice Sci, 31(6): 665-672. (in Chinese with English abstract) |
| [10] | Huang R Y, Jiang L R, Zheng J S, Wang T S, Wang H C, Huang Y M, Hong Z L.2013. Genetic bases of rice grain shape: So many genes, so little known.Trends Plant Sci, 18(4): 218-226. |
| [11] | Ishimaru K.2003. Identification of a locus increasing rice yield and physiological analysis of its function.Plant Physiol, 133(3): 1083-1090. |
| [12] | Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, Miyagawa H, Katoh E.2013. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet, 45(6): 707-711. |
| [13] | Li Q, Li L, Yang X H, Warburton M L, Bai G H, Dai J R, Li J S, Yan J B.2010a. Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol, 10(1): 143. |
| [14] | Li Q, Yang X H, Bai G H, Warburtion M L, Mahuku G, Gore M, Dai J R, Li J S, Yan J B.2010b. Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor Appl Genet, 120(4): 753-763. |
| [15] | Li Y B, Fan C C, Xing Y Z, Jiang Y H, Luo L J, Sun L, Shao D, Xu C J, Li X H, Xiao J H, He Y Q, Zhang Q F.2011. Natural variation inGS5 plays an important role in regulating grain size and yield in rice. Nat Genet, 43(12): 1266-1269. |
| [16] | Li Y B, Fan C C, Xing Y Z, Yun P, Luo L J, Yan B, Peng B, Xie W B, Wang G W, Li X H, Xiao J H, Xu C G, He Y Q.2014. Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice.Nat Genet, 46(4): 398-404. |
| [17] | Meyer R S, Purugganan M D.2013. Evolution of crop species: Genetics of domestication and diversification.Nat Rev Genet, 14(12): 840-852. |
| [18] | Mignone F, Gissi C, Liuni S, Pesole G.2002. Untranslated regions of mRNAs.Genome Biol, 3(3): 1-10. |
| [19] | Moles A T, Ackerly D D, Webb C O, Tweddle J C, Dickie J B, Westoby M.2005. A brief history of seed size.Science, 307: 576-580. |
| [20] | Sakamoto T, Matsuoka M.2008. Identifying and exploiting grain yield genes in rice.Curr Opin Plant Biol, 11(2): 209-214. |
| [21] | Scofield G N, Hirose T, Aoki N, Furbank R T.2007. Involvement of the sucrose transporter,OsSUT1, in the long-distance pathway for assimilate transport in rice. J Exp Bot, 58(12): 3155-3169. |
| [22] | She K C, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M, Tsuge T, Matsumoto K, Kudoh M, Itoh E, Kikuchi S, Kishimoto N, Yazaki J, Ando T, Yano M, Aoyama T, Sasaki T, Satoh H, Shimada H.2010. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell, 22(10): 3280-3294. |
| [23] | Si L Z, Chen J Y, Huang X H, Gong H, Luo J H, Hou Q Q, Zhou T Y, Lu T T, Zhu J J, Shangguan Y Y, Chen E W, Gong C X, Zhao Q, Jing Y F, Zhao Y, Li Y, Cui L L, Fan D L, Lu Y Q, Weng Q J, Wang Y C, Zhan Q L, Liu K Y, Wei X H, An K, An G, Han B.2016. OsSPL13 controls grain size in cultivated rice.Nat Genet, 48(4): 447-456. |
| [24] | Song X J, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, Suzuki T, Higashiyama T, Yamasaki M, Mori H, Inukai Y, Wu J Z, Kitano H, Sakakibara H, Jacobsen S E, Ashikari M.2015. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice.Prol Natl Acad Sci USA, 112(1): 76-81. |
| [25] | Sreenivasulu N, Butardo V M, Misra G, Cuevas R P, Anacleto R, Kishor P B K.2015. Designing climate-resilient rice with ideal grain quality suited for high-temperature stress.J Exp Bot, 66(7): 1737-1748. |
| [26] | Su Y, Rao Y C, Hu S K, Yang Y L, Gao Z Y, Zhang G H, Liu J, Hu J, Yan M X, Dong G J, Zhu L, Guo L B, Qian Q, Zeng D L.2011. Map-based cloning proves qGC-6, a major QTL for gel consistency of japonica/indica cross, responds by Waxy in rice (Oryza sativa L.). Theor Appl Genet, 123(5): 859-867. |
| [27] | Sun M M, Abdula S E, Lee H J, Cho Y C, Han L Z, Koh H J, Cho Y G.2011. Molecular aspect of good eating quality formation in japonica rice. PLoS One, 6(4): e18385. |
| [28] | Sweeney M, McCouch S.2007. The complex history of the domestication of rice.Ann Bot-London, 100(5): 951-957. |
| [29] | Takano-Kai N, Jiang H, Kubo T, Sweeney M, Matsumoto T, Kanamori H, Padhukasahasram B, Bustamante C, Yoshimura A,Doi K, McCouch S. 2009. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics, 182(4): 1323-1334. |
| [30] | Takano-Kai N, Doi K, Yoshimura A.2011. GS3 participates in stigma exsertion as well as seed length in rice.Breeding Sci, 61(3): 244-250. |
| [31] | Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, Yano M, Yoshimura A, Kitano H, Matsuoka M, Fujisawa Y, Kato H, Iwasaki Y.2005. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant,dwarf11, with reduced seed length. Plant Cell, 17(3): 776-790. |
| [32] | Velden A W V D, Thomas A A M.1999. The role of the 5′ untranslated region of an mRNA in translation regulation during development.Int J Biochem Cell B, 31(1): 87-106. |
| [33] | Wang L Q, Liu W J, Xu Y, He Y Q, Luo L J, Xing Y Z, Xu C G, Zhang Q F.2007. Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of rice grain.Theor Appl Genet, 115(4): 463-476. |
| [34] | Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D.2012. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet, 44(8): 950-954. |
| [35] | Wang S K, Li S, Liu Q, Wu K, Zhang J Q, Wang S S, Wang Y, Chen X B, Zhang Y, Gao C X, Wang F, Huang H X, Fu X D.2015. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet, 47(8): 949-954. |
| [36] | Wang Y X, Xiong G S, Hu J, Jiang L, Yu H, Xu J, Fang Y X, Zeng L J, Xu E B, Xu J, Ye W J, Meng X B, Liu R F, Chen H Q, Jing Y H, Wang Y H, Zhu X D, Li J Y, Qian Q.2015. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet, 47(8): 944-948. |
| [37] | Xing Y Z, Zhang Q F.2010. Genetic and molecular bases of rice yield.Annu Rev Plant Biol, 61: 421-442. |
| [38] | Yi C D, Wang D R, Jiang W, Li W, Cheng X J, Wang Y, Zhou Y, Liang G H, Gu M H.2016. Development of functional markers and identification of haplotypes for rice grain width gene GS5. Chin J Rice Sci, 30(5): 487-492. (in Chinese with English abstract) |
| [39] | Zhu M Z, Shi M, Lin H X, Huang W, Song X J.2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase.Nat Genet, 39(5): 623-630. |
| [40] | Zuo J R, Li J Y.2014. Molecular genetic dissection of quantitative trait loci regulating rice grain size.Annu Rev Genet, 48: 99-118. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||