
Rice Science ›› 2018, Vol. 25 ›› Issue (3): 152-1604.DOI: 10.1016/j.rsci.2018.04.003
收稿日期:2018-01-04
接受日期:2018-02-26
出版日期:2018-05-04
发布日期:2018-03-07
. [J]. Rice Science, 2018, 25(3): 152-1604.
| Gene | Accession | Gene description | Forward primer (5′-3′) | Reverse primer (5′-3′) | Production size (bp) |
|---|---|---|---|---|---|
| OsALDH2-1 | AK119571 | Aldehyde dehydrogenase 2-1 | CCTCAGATTGACAAGGAGCA | GGCTCGATGTAGTACCCGTT | 122 |
| OsALDH2-2 | AK101427 | Aldehyde dehydrogenase 2-2 | GGTCCTCAGGTTGACAAGGT | CTTTGTCACCAGTGGGTTTG | 109 |
| OsALDH2-3 | CU607043 | Aldehyde dehydrogenase 2-3 | CAGGTGGACAAGGCTCAGTA | AATGGTGGGCTCGATGTAGT | 126 |
| OsALDH2-4 | AK121610 | Aldehyde dehydrogenase 2-4 | TCTTGCCTGGATCACTTCAC | CATTGATGAGGAGCTTGGTG | 120 |
| OsALDH2-5 | AK073079 | Aldehyde dehydrogenase 2-5 | AAGTCGTCCTTGAGTTGGCT | CATGGTCAACATCAGCATCA | 105 |
| OsALDH3-1 | AK070741 | Aldehyde dehydrogenase 3-1 | ATAATCGGAGCGAAATGGTC | CAATCAGGAATGGTGCAAAC | 94 |
| OsALDH3-2 | AK120274 | Aldehyde dehydrogenase 3-2 | AGGGTGGCAGCTTCTGTAGT | TTACCGTGATGATTGGGAGA | 145 |
| OsALDH3-3 | AK071169 | Aldehyde dehydrogenase 3-3 | ATAGAGGACAGCATCGCCTT | ACTGCGTCGTTGAACGTAAC | 134 |
| OsALDH3-4 | AK104746 | Aldehyde dehydrogenase 3-4 | ATTCCCATCAACTGCACAAA | TATGACTGGGTCGATGGAGA | 100 |
| OsALDH3-5 | JC625236 | Aldehyde dehydrogenase 3-5 | GCTGCGATCATCTGACTTGT | TCGTCCATCAGCTTCTTCAG | 72 |
| OsALDH7 | AK120185 | Aldehyde dehydrogenase 7 | AGAGCAAAGCTCCATCACCT | TGAACCTCTCCAATCCCTTC | 80 |
| OsAKR1 | AK242899 | Aldo-keto reductase 1 | TCGATTGCATCGACCTTTAC | ATGCCGATGCTTCACATAAG | 132 |
| OsAKR2 | AK103729 | Aldo-keto reductase 2 | CATCAGGGAAGCGATGTATG | CCTCGAACATCATCTGTGCT | 143 |
| OsAKR3 | AK100718 | Aldo-keto reductase 3 | AGCTCACACCTGACGAGATG | GCGGAGGAGTTTCAGAGTTC | 117 |
| UBQ5 | AK062354 | Ubiquitin 5 | ACCACTTCGACCGCCACTACT | ACGCC TAAGCCTGCTGGTT | 69 |
Table 1 Primers used in real-time PCR.
| Gene | Accession | Gene description | Forward primer (5′-3′) | Reverse primer (5′-3′) | Production size (bp) |
|---|---|---|---|---|---|
| OsALDH2-1 | AK119571 | Aldehyde dehydrogenase 2-1 | CCTCAGATTGACAAGGAGCA | GGCTCGATGTAGTACCCGTT | 122 |
| OsALDH2-2 | AK101427 | Aldehyde dehydrogenase 2-2 | GGTCCTCAGGTTGACAAGGT | CTTTGTCACCAGTGGGTTTG | 109 |
| OsALDH2-3 | CU607043 | Aldehyde dehydrogenase 2-3 | CAGGTGGACAAGGCTCAGTA | AATGGTGGGCTCGATGTAGT | 126 |
| OsALDH2-4 | AK121610 | Aldehyde dehydrogenase 2-4 | TCTTGCCTGGATCACTTCAC | CATTGATGAGGAGCTTGGTG | 120 |
| OsALDH2-5 | AK073079 | Aldehyde dehydrogenase 2-5 | AAGTCGTCCTTGAGTTGGCT | CATGGTCAACATCAGCATCA | 105 |
| OsALDH3-1 | AK070741 | Aldehyde dehydrogenase 3-1 | ATAATCGGAGCGAAATGGTC | CAATCAGGAATGGTGCAAAC | 94 |
| OsALDH3-2 | AK120274 | Aldehyde dehydrogenase 3-2 | AGGGTGGCAGCTTCTGTAGT | TTACCGTGATGATTGGGAGA | 145 |
| OsALDH3-3 | AK071169 | Aldehyde dehydrogenase 3-3 | ATAGAGGACAGCATCGCCTT | ACTGCGTCGTTGAACGTAAC | 134 |
| OsALDH3-4 | AK104746 | Aldehyde dehydrogenase 3-4 | ATTCCCATCAACTGCACAAA | TATGACTGGGTCGATGGAGA | 100 |
| OsALDH3-5 | JC625236 | Aldehyde dehydrogenase 3-5 | GCTGCGATCATCTGACTTGT | TCGTCCATCAGCTTCTTCAG | 72 |
| OsALDH7 | AK120185 | Aldehyde dehydrogenase 7 | AGAGCAAAGCTCCATCACCT | TGAACCTCTCCAATCCCTTC | 80 |
| OsAKR1 | AK242899 | Aldo-keto reductase 1 | TCGATTGCATCGACCTTTAC | ATGCCGATGCTTCACATAAG | 132 |
| OsAKR2 | AK103729 | Aldo-keto reductase 2 | CATCAGGGAAGCGATGTATG | CCTCGAACATCATCTGTGCT | 143 |
| OsAKR3 | AK100718 | Aldo-keto reductase 3 | AGCTCACACCTGACGAGATG | GCGGAGGAGTTTCAGAGTTC | 117 |
| UBQ5 | AK062354 | Ubiquitin 5 | ACCACTTCGACCGCCACTACT | ACGCC TAAGCCTGCTGGTT | 69 |
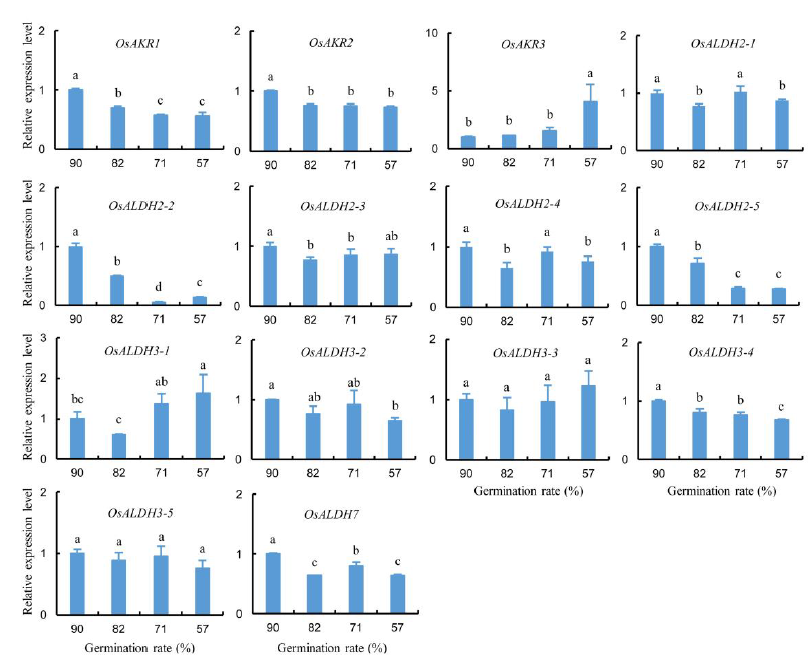
Fig. 4. Expression analysis of ALDH and AKR genes in aged rice embryos. UBQ5 gene was used as a housekeeping control. Data represent Mean ± SD. Seeds with the germination rate of 90% were served as the control. Different letters mean significant difference among treatments (P < 0.05, one-way ANOVA, n = 3).
| [1] | Alméras E, Stolz S, Vollenweider S, Reymond P, Mène-Saffrané L, Farmer E E.2003. Reactive electrophile species activate defense gene expression inArabidopsis. Plant J, 34(2): 205-216. |
| [2] | Csallany A S, Han I, Shoeman D W, Chen C, Yuan J Y.2015. 4-hydroxynonenal (HNE), a toxic aldehyde in French fries from fast food restaurants.J Am Oil Chem Soc, 92(10): 1413-1419. |
| [3] | FAO.2010. Second report on the state of the world’s plants genetic resources for food and agriculture. Commission on Genetic Resources for Food and Agriculture. Rome: Food and Agriculture Organization of the United Nations: 47. |
| [4] | Farmer E E, Mueller M J.2013. ROS-mediated lipid peroxidation and RES-activated signaling.Annu Rev Plant Biol, 64(1): 429-450. |
| [5] | Gao C X, Han B.2009. Evolutionary and expression study of the aldehyde dehydrogenase (ALDH) gene superfamily in rice (Oryza sativa). Gene, 431(1/2): 86-94. |
| [6] | Gao H Y, Jing L Q, Chen L, Ju J, Wang Y X, Zhu J G, Yang L X, Wang Y L.2016. Effects of elevated atmospheric CO2 and temperature on seed vigor of rice under open-air field conditions.Chin J Rice Sci, 30(4): 371-379. (in Chinese with English abstract) |
| [7] | Hay F R, de Guzman F, Ellis D, Makahiya H, Borromeo T, Hamilton N R S.2013. Viability of Oryza sativa L. seeds stored under genebank conditions for up to 30 years. Genet Resour Crop Evol, 60(1): 275-296. |
| [8] | Hay F R, de Guzman F, Hamilton N R S.2015. Viability monitoring intervals for genebank samples ofOryza sativa. Seed Sci Technol, 43(2): 218-237. |
| [9] | Hideg É, Nagy T, Oberschall A, Dudits D, Vass I.2003. Detoxification function of aldose/aldehyde reductase during drought and ultraviolet-B (280-320 nm) stresses.Plant Cell Environ, 26(4): 513-522. |
| [10] | Islam M M, Ye W, Matsushima D, Munemasa S, Okuma E, Nakamura Y, Biswas S, Mano J, Murata Y.2016. Reactive carbonyl species mediate ABA signaling in guard cells.Plant Cell Physiol, 57(12): 2552-2563. |
| [11] | ISTA.2014. International Rules for Seed Testing. Bassersdorf, Switzerland: International Seed Testing Association. |
| [12] | Kotchoni S O, Kuhns C, Ditzer A, Kirch H H, Bartels D.2006. Overexpression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ, 29(6): 1033-1048. |
| [13] | Lu X X, Chen X L, Guo Y H.2005. Seed germinability of 23 crop species after a decade of storage in the national genebank of China.Agric Sci China, 4(6): 408-412. |
| [14] | Mano J, Miyatake F, Hiraoka E, Tamoi M.2009. Evaluation of the toxicity of stress-related aldehydes to photosynthesis in chloroplasts.Planta, 230(4): 639-648. |
| [15] | Mano J.2012. Reactive carbonyl species: Their production from lipid peroxides, action in environmental stress, and the detoxification mechanism.Plant Physiol Biochem, 59: 90-97. |
| [16] | Mano J, Nagata M, Okamura S, Shiraya T, Mitsui T.2014. Identification of oxidatively modified proteins in salt-stressed Arabidopsis: A carbonyl-targeted proteomics approach. Plant Cell Physiol, 55(7): 1233-1244. |
| [17] | Matsui K, Sugimoto K, Kakumyan P, Khorobrykh S A, Mano J.2009. Volatile oxylipins and related compounds formed under stress in plants.Methods Mol Biol, 580: 17-28. |
| [18] | Matsui K, Sugimoto K, Mano J, Ozawa R, Takabayashi J.2012. Differential metabolisms of green leaf volatiles in injured and intact parts of a wounded leaf meet distinct ecophysiological requirements.PLoS One, 7(4): e36433. |
| [19] | Mène-Saffrané L, Davoine C, Stolz S, Majcherczyk P, Farmer E E.2007. Genetic removal of tri-unsaturated fatty acids suppresses developmental and molecular phenotypes of an Arabidopsis tocopherol-deficient mutant: Whole-body mapping of malondialdehyde pools in a complex eukaryote. J Biol Chem, 282: 35749-35756. |
| [20] | Mueller M J.1998. Radically novel prostaglandins in animals and plants: The isoprostanes.Chem Biol, 5(12): 323-333. |
| [21] | Nakazono M, Tsuji H, Li Y, Saisho D, Arimura S, Tsutsumi N, Hirai A.2000. Expression of a gene encoding mitochondrial aldehyde dehydrogenase in rice increases under submerged conditions.Plant Physiol, 124(2): 587-598. |
| [22] | Oberschall A, Deák M, Török K, Sass L, Vass I, Kovács I, Fehér A, Dudits D, Horváth G V.2000. A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses.Plant J, 24(4): 437-446. |
| [23] | Penning T M.2015. The aldo-keto reductases (AKRs): Overview.Chem-Biol Inter, 234(5): 236-246. |
| [24] | Rodrigues S M, Andrade M O, Gomes A P S, Damatta F M, Baracat-Pereira M C, Fontes E P B.2006. Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J Exp Bot, 57(9): 1909-1918. |
| [25] | Sengupta D, Naik D, Reddy A R.2015. Plant aldo-keto reductases (AKRs) as multi-tasking soldiers involved in diverse plant metabolic processes and stress defense: A structure-function update.J Plant Physiol, 179: 40-55. |
| [26] | Shin J H, Kim S R, An G.2009. Rice aldehyde dehydrogenase 7 is needed for seed maturation and viability.Plant Physiol, 149(2): 905-915. |
| [27] | Stiti N, Adewale I O, Petersen J, Bartels D, Kirch H H.2011. Engineering the nucleotide coenzyme specificity and sulfhydryl redox sensitivity of two stress-responsive aldehyde dehydrogenase isoenzymes of Arabidopsis thaliana. Biochem J, 434(3): 459-471. |
| [28] | Sun Y L, Liu H M, Xu Q G.2017. Effects of cadmium stress on rice seed germination characteristics.Chin J Rice Sci, 31(4): 425-431. (in Chinese with English abstract) |
| [29] | Sunkar R, Bartels D, Kirch H H.2003. Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J, 35(4): 452-464. |
| [30] | Taylor N L, Day D A, Millar A H.2002. Environmental stress causes oxidative damage to plant mitochondria leading to inhibition of glycine decarboxylase.J Biol Chem, 277(45): 42663-42668. |
| [31] | Tsuji H, Meguro N, Suzuki Y, Tsutsumi N, Hirai A, Nakazono M.2003. Induction of mitochondrial aldehyde dehydrogenase by submergence facilitates oxidation of acetaldehyde during re-aeration in rice.Febs Lett, 546: 369-373. |
| [32] | Turóczy Z, Kis P, Török K, Cserháti M, Lendvai Á, Dudits D, Horváth G V.2011. Overproduction of a rice aldo-keto reductase increases oxidative and heat stress tolerance by malondialdehyde and methylglyoxal detoxification.Plant Mol Biol, 75(4/5): 399-412. |
| [33] | Vemanna R S, Vennapusa A R, Easwaran M, Chandrashekar B K, Rao H, Ghanti K, Sudhakar C, Mysore K S, Makarla U.2016. Aldo-keto reductase enzymes detoxify glyphosate and improve herbicide resistance in plants.Plant Biotechnol J, 15(7): 794-804. |
| [34] | Vemanna R S, Babitha K C, Solanki J K, Amarnatha Reddy V, Sarangi S K, Udayakumar M.2017. Aldo-keto reductase-1 (AKR1) protect cellular enzymes from salt stress by detoxifying reactive cytotoxic compounds.Plant Physiol Biochem, 113: 177-186. |
| [35] | Wang H, Jiang Z Z, Du H B, Liang C Y, Wang Y C, Zhang M H, Zhang L Y, Ye W C, Li P.2012. Simultaneous determination of three flavonoid C-glycosides in mice biosamples by HPLC- ESI-MS method after oral administration of Abrus mollis extract and its application to biodistribution studies. J Chromatogr B, 903: 68-74. |
| [36] | Weber H, Chételat A, Reymond P, Farmer E E.2004. Selective and powerful stress gene expression inArabidopsis in response to malondialdehyde. Plant J, 37(6): 877-888. |
| [37] | Xin X, Lin X H, Zhou Y C, Chen X L, Liu X, Lu X X.2011. Proteome analysis of maize seeds: The effect of artificial ageing.Physiol Planta, 143(2): 126-138. |
| [38] | Yamauchi Y, Hasegawa A, Taninaka A, Mizutani M, Sugimoto Y.2011. NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants.J Biol Chem, 286(9): 6999-7009. |
| [39] | Yin G K, Whelan J, Wu S H, Zhou J, Chen X L, Chen B Y, Zhang J M, Xin X, Lu X X.2016. Comprehensive mitochondrial metabolic shift during the critical node of seed ageing in rice.PLoS One, 11(4): e0148013. |
| [40] | Yin G K, Xin X, Fu S Z, An M N, Wu S H, Chen X L, Zhang J M, He J J, Whelan J, Lu X X.2017. Proteomic and carbonylation profile analysis at the critical node of seed ageing inOryza sativa. Sci Rep-UK, 7: 40611. |
| [41] | Yin L, Mano J, Wang S, Tsuji W, Tanaka K.2010. The involvement of lipid peroxide-derived aldehydes in aluminum toxicity of tobacco roots.Plant Physiol, 152(3): 1406-1417. |
| [42] | Zhou J L, Wang X F, Jiao Y L, Qin Y H, Liu X G, He K, Chen C, Ma L G, Wang J, Xiong L Z, Zhang Q F, Fan L M, Deng X W.2007. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle.Plant Mol Biol, 63(5): 591-608. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||