
Rice Science ›› 2024, Vol. 31 ›› Issue (6): 700-711.DOI: 10.1016/j.rsci.2024.08.004
• Research Papers • Previous Articles Next Articles
Li Wei1,#, Zhang Mengchen2,#, Yang Yaolong2, Weng Lin1, Hu Peisong2( ), Wei Xinghua3(
), Wei Xinghua3( )
)
Received:2024-05-27
Accepted:2024-08-26
Online:2024-11-28
Published:2024-12-10
Contact:
Wei Xinghua (weixinghua@caas.cn);
Hu Peisong (peisonghu@126.com)
About author:#These authors contributed equally to this work
Li Wei, Zhang Mengchen, Yang Yaolong, Weng Lin, Hu Peisong, Wei Xinghua. Molecular Evolution of Rice Blast Resistance Gene bsr-d1[J]. Rice Science, 2024, 31(6): 700-711.
Add to citation manager EndNote|Ris|BibTeX
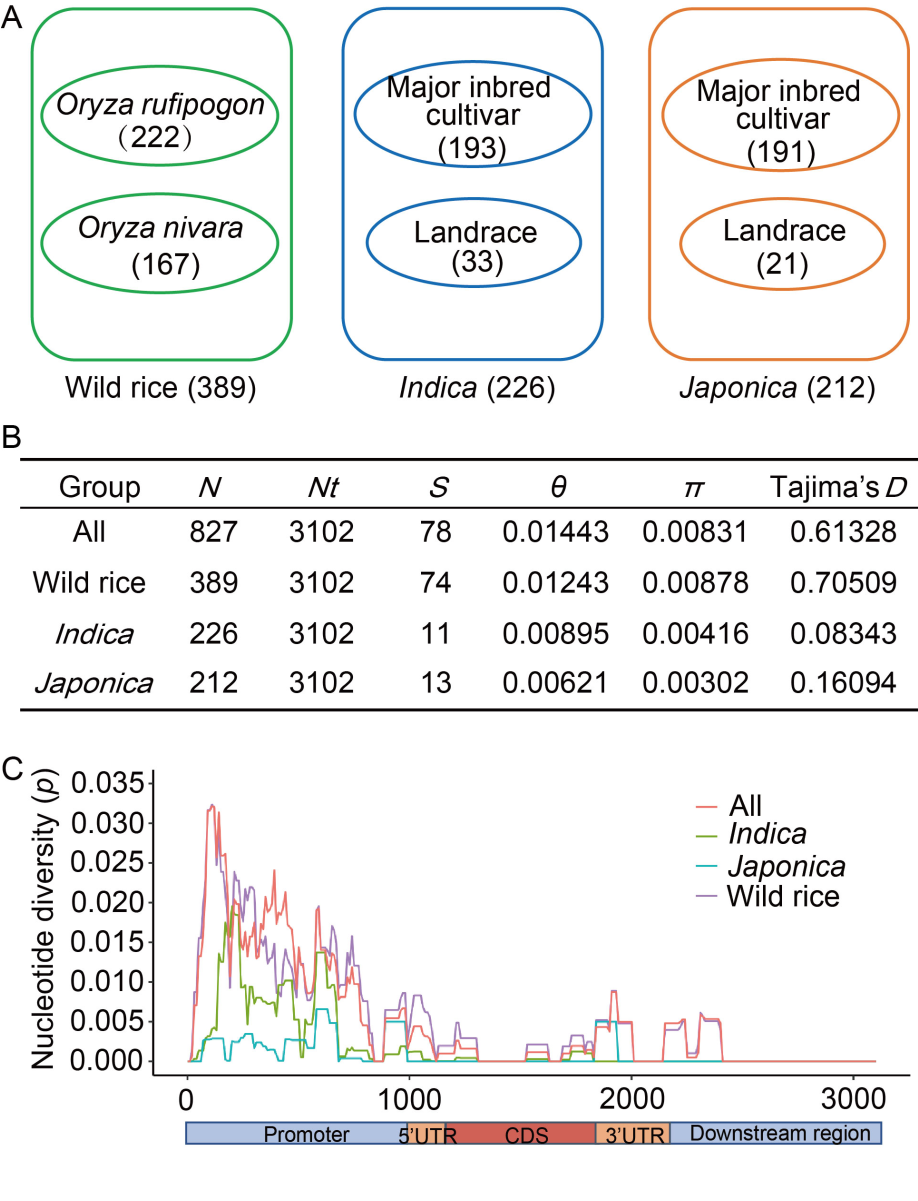
Fig. 1. Nucleotide polymorphisms of Bsr-d1 in japonica, indica, and wild rice. A, Composition of 827 rice accessions used for analysis. B, Nucleotide diversity of Bsr-d1 in japonica, indica, and wild rice. N indicates sample size; Nt indicates the number of sites in the analyzed sequence; S indicates the number of polymorphic sites; θ indicates silent nucleotide polymorphism; π indicates nucleotide diversity based on silent sites; Tajima’s D statistic is based on the differences between the number of segregating sites and the average number of nucleotide differences. C, Sliding window of nucleotide diversity (p) of Bsr-d1 in three subgroups. The X-axis represents the nucleotide position along Bsr-d1, and the Y-axis indicates the value of nucleotide diversity per site. The sliding window is 100 bp with a step size of 10 bp. The promoter, 5ʹ UTR (untranslated region), CDS (coding sequence), 3ʹ UTR, and downstream regions are marked on the corresponding region below the sliding windows in their respective areas.
| Group | Component | S | θ | π | Tajima’s D |
|---|---|---|---|---|---|
| All | Promoter | 63 | 0.02262 | 0.01376 | 0.96874 |
| 5ʹ UTR | 3 | 0.01287 | 0.00284 | 0.51108 | |
| CDS | 5 | 0.00437 | 0.00142 | 0.04525 | |
| 3ʹ UTR | 2 | 0.00547 | 0.00472 | 1.02496 | |
| Downstream region | 5 | 0.00679 | 0.00410 | 0.85622 | |
| Wild rice | Promoter | 60 | 0.02262 | 0.01387 | 0.94544 |
| 5ʹ UTR | 3 | 0.01359 | 0.00572 | 0.12206 | |
| CDS | 5 | 0.00437 | 0.00257 | 0.36591 | |
| 3ʹ UTR | 1 | 0.00547 | 0.00507 | 0.76849 | |
| Downstream region | 5 | 0.00679 | 0.00389 | 0.61085 | |
| Indica | Promoter | 10 | 0.01124 | 0.00624 | 0.15972 |
| 5ʹ UTR | 0 | 0.00000 | 0.00000 | 0.00000 | |
| CDS | 1 | 0.00425 | 0.00049 | 0.54851 | |
| 3ʹ UTR | 0 | 0.00000 | 0.00000 | 0.00000 | |
| Downstream region | 0 | 0.00000 | 0.00000 | 0.00000 | |
| Japonica | Promoter | 12 | 0.00654 | 0.00278 | 0.09672 |
| 5ʹ UTR | 0 | 0.00000 | 0.00000 | 0.00000 | |
| CDS | 0 | 0.00000 | 0.00000 | 0.00000 | |
| 3ʹ UTR | 1 | 0.00353 | 0.00104 | 1.04530 | |
| Downstream region | 0 | 0.00000 | 0.00000 | 0.00000 |
Table 1. Nucleotide polymorphisms of Bsr-d1 in different gene regions.
| Group | Component | S | θ | π | Tajima’s D |
|---|---|---|---|---|---|
| All | Promoter | 63 | 0.02262 | 0.01376 | 0.96874 |
| 5ʹ UTR | 3 | 0.01287 | 0.00284 | 0.51108 | |
| CDS | 5 | 0.00437 | 0.00142 | 0.04525 | |
| 3ʹ UTR | 2 | 0.00547 | 0.00472 | 1.02496 | |
| Downstream region | 5 | 0.00679 | 0.00410 | 0.85622 | |
| Wild rice | Promoter | 60 | 0.02262 | 0.01387 | 0.94544 |
| 5ʹ UTR | 3 | 0.01359 | 0.00572 | 0.12206 | |
| CDS | 5 | 0.00437 | 0.00257 | 0.36591 | |
| 3ʹ UTR | 1 | 0.00547 | 0.00507 | 0.76849 | |
| Downstream region | 5 | 0.00679 | 0.00389 | 0.61085 | |
| Indica | Promoter | 10 | 0.01124 | 0.00624 | 0.15972 |
| 5ʹ UTR | 0 | 0.00000 | 0.00000 | 0.00000 | |
| CDS | 1 | 0.00425 | 0.00049 | 0.54851 | |
| 3ʹ UTR | 0 | 0.00000 | 0.00000 | 0.00000 | |
| Downstream region | 0 | 0.00000 | 0.00000 | 0.00000 | |
| Japonica | Promoter | 12 | 0.00654 | 0.00278 | 0.09672 |
| 5ʹ UTR | 0 | 0.00000 | 0.00000 | 0.00000 | |
| CDS | 0 | 0.00000 | 0.00000 | 0.00000 | |
| 3ʹ UTR | 1 | 0.00353 | 0.00104 | 1.04530 | |
| Downstream region | 0 | 0.00000 | 0.00000 | 0.00000 |
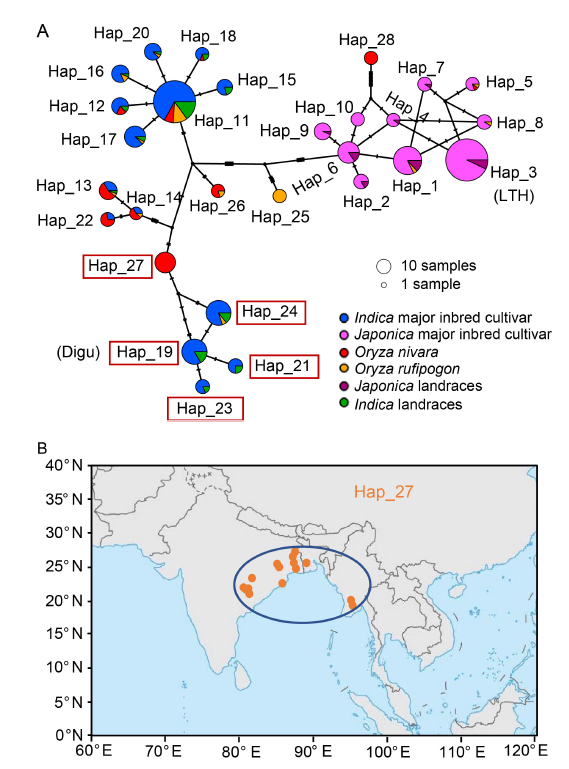
Fig. 2. Origin and evolution of Bsr-d1 haplotypes. A, Haplotype network of Bsr-d1. The size of each circle is proportional to the number of samples for a given haplotype. Red boxes represent resistance haplotypes. The highly susceptible rice variety Lijiangxintuanheigu (LTH) is classified under Hap_3, while the highly resistant rice variety Digu is classified under Hap_19. B, Geographic distribution of Oryza nivara belonging to Hap_27. Orange dots represent the geographical distribution of Hap_27. The area enclosed by the blue ellipse highlights the region adjacent to the the Indian Peninsula and the Indochina Peninsula.
| Haplotype | Sequence |
|---|---|
| Reference | TCAGTTGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_1 | TCAGTCGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_2 | TCAGTTGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCACGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_3 | TCAGTCGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCACCTCGGGTGCTCCCGCGCGGA |
| Hap_4 | TCAGTCGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCACGGGGCCAACTCGGGTGCTCCCGCGCGGA |
| Hap_5 | TCAGTCGAGCTAGCCTGGGATGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCACCTCGGGTGCTCCCGCGCGGA |
| Hap_6 | TCAGTCGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCACGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_7 | TCAGTCGAGCTAGCCTGGGATGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCACCTCGGGTGCTCCCACGCGGA |
| Hap_8 | TCAGTCGAGCTAGCCTGGGATGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCAACTCGGGTGCTCCCGCGCGGA |
| Hap_9 | TCAGTCGAGCTAGCCTGGGATGCGACCAAGGCTCCCTGGTGCCATGCCACGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_10 | TCAGTCGAGCTAGCCTGGGATGCGACCAAGGCTCCCTGGCGCCACGCCACGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_11 | TAAGACGCGTTCGCCTGAGATGCGACCGAGGACCCTCGACGCTATGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_12 | TAAGACGCGTTCGCCTGAGATGCGACCGAGGACCCTCGACACTATGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_13 | TAACACGCGTTCGCCCGAGATGCGACCGAGGACCCCCGACACTATGTCTTGGGCCAACCCCGGGTGCTCTCGTACGGC |
| Hap_14 | TAACACGCGTTCGCCCGAGATGCGACCGAGGACCCCCGACACTATGTCTCGGGCCAACCCCGGGTGCTCTCGTACGGC |
| Hap_15 | TAAGACGCGTTCGCCTGAGATGCGACCGAGGACCCTCGACGCTATGCCTCGGGGCCAACTCGGGTGCTCCCGTACGGC |
| Hap_16 | TAAGACGCTTTGGCCTGAGATGCGACCGAGGACCCTCGACGCTATGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_17 | TAAGACGCGTTCGCCTGAGATGCGACCGAGGACCCTCGACGCTATGCCTTGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_18 | TAAGACGCGTTCGCCTGAGATGCGACCGAGGACCCTCGACGCTATGCCTCGGCCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_19 | TAAGACGCGTTCGCTCGGGATGCGATCGAGGACCCCCTACGCTGTGCGTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_20 | TAAGACGCGTTCCCCTGAGGTGCGACCGAGGACCCTCGACGCTATGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_21 | TAAGACGCGTTCGCTCGGGATGCGATCGAGGACCCCCTACGCTGTGTGTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_22 | TAACACGCGTTCGTCCGGGGTGCGACCGAGGACCCCCGACGCTATGTCTCGGGCCATCCCCGGGTGCTCTCGTACGGC |
| Hap_23 | TAAGACGCGTTCGCTCGGGATGCGATCGGGGACCCCCTACGCTGTGCGTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_24 | TAAGACGCGTTCGCTCGGGGTGCGATCGAGGACCCCCTACGCTGTGCGTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_25 | TATGACACGCTAGTCTGGGGTGCGACCGAGAACTCCCGACGCCATGCCACGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_26 | TAAGACGCGTTCGCTTGGGATGCGACCGAGGACCCCCGACGCTATGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_27 | TAAGACGCGTTCGCCCGGGGTGCGATCGAGGACCCCCGACGCTGTGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_28 | TCAGTCGCTCTAGCCTGGGACACGACCGAGGACCCTCGGCGCCACGCCACGGGGACAACTCGGGTGCTCCCGCGCGGA |
Table 2. Nucleotide information of 28 haplotypes of Bsr-d1.
| Haplotype | Sequence |
|---|---|
| Reference | TCAGTTGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_1 | TCAGTCGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_2 | TCAGTTGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCACGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_3 | TCAGTCGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCACCTCGGGTGCTCCCGCGCGGA |
| Hap_4 | TCAGTCGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCACGGGGCCAACTCGGGTGCTCCCGCGCGGA |
| Hap_5 | TCAGTCGAGCTAGCCTGGGATGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCACCTCGGGTGCTCCCGCGCGGA |
| Hap_6 | TCAGTCGAGCTAGCCTGGGGTGCGACCAAGGCTCCCTGGCGCCATGCCACGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_7 | TCAGTCGAGCTAGCCTGGGATGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCACCTCGGGTGCTCCCACGCGGA |
| Hap_8 | TCAGTCGAGCTAGCCTGGGATGCGACCAAGGCTCCCTGGCGCCATGCCTCGGGGCCAACTCGGGTGCTCCCGCGCGGA |
| Hap_9 | TCAGTCGAGCTAGCCTGGGATGCGACCAAGGCTCCCTGGTGCCATGCCACGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_10 | TCAGTCGAGCTAGCCTGGGATGCGACCAAGGCTCCCTGGCGCCACGCCACGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_11 | TAAGACGCGTTCGCCTGAGATGCGACCGAGGACCCTCGACGCTATGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_12 | TAAGACGCGTTCGCCTGAGATGCGACCGAGGACCCTCGACACTATGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_13 | TAACACGCGTTCGCCCGAGATGCGACCGAGGACCCCCGACACTATGTCTTGGGCCAACCCCGGGTGCTCTCGTACGGC |
| Hap_14 | TAACACGCGTTCGCCCGAGATGCGACCGAGGACCCCCGACACTATGTCTCGGGCCAACCCCGGGTGCTCTCGTACGGC |
| Hap_15 | TAAGACGCGTTCGCCTGAGATGCGACCGAGGACCCTCGACGCTATGCCTCGGGGCCAACTCGGGTGCTCCCGTACGGC |
| Hap_16 | TAAGACGCTTTGGCCTGAGATGCGACCGAGGACCCTCGACGCTATGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_17 | TAAGACGCGTTCGCCTGAGATGCGACCGAGGACCCTCGACGCTATGCCTTGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_18 | TAAGACGCGTTCGCCTGAGATGCGACCGAGGACCCTCGACGCTATGCCTCGGCCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_19 | TAAGACGCGTTCGCTCGGGATGCGATCGAGGACCCCCTACGCTGTGCGTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_20 | TAAGACGCGTTCCCCTGAGGTGCGACCGAGGACCCTCGACGCTATGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_21 | TAAGACGCGTTCGCTCGGGATGCGATCGAGGACCCCCTACGCTGTGTGTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_22 | TAACACGCGTTCGTCCGGGGTGCGACCGAGGACCCCCGACGCTATGTCTCGGGCCATCCCCGGGTGCTCTCGTACGGC |
| Hap_23 | TAAGACGCGTTCGCTCGGGATGCGATCGGGGACCCCCTACGCTGTGCGTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_24 | TAAGACGCGTTCGCTCGGGGTGCGATCGAGGACCCCCTACGCTGTGCGTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_25 | TATGACACGCTAGTCTGGGGTGCGACCGAGAACTCCCGACGCCATGCCACGGGGCCAACTCGGGTGCTCCCACGCGGA |
| Hap_26 | TAAGACGCGTTCGCTTGGGATGCGACCGAGGACCCCCGACGCTATGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_27 | TAAGACGCGTTCGCCCGGGGTGCGATCGAGGACCCCCGACGCTGTGCCTCGGGCCCACCTCGGGTGCTCCCGTACGGC |
| Hap_28 | TCAGTCGCTCTAGCCTGGGACACGACCGAGGACCCTCGGCGCCACGCCACGGGGACAACTCGGGTGCTCCCGCGCGGA |
| Group | N | Nt | S | θ | π | Tajima’s D |
|---|---|---|---|---|---|---|
| Oryza nivara | 167 | 3 102 | 74 | 0.01323 | 0.00896 | 0.76345 |
| Indica landrace | 33 | 3 102 | 11 | 0.00946 | 0.00543 | 0.14335 |
| Indica major inbred cultivar | 193 | 3 102 | 9 | 0.00621 | 0.00354 | 0.08786 |
Table 3. Nucleotide diversity of Bsr-d1 in Oryza nivara, indica landraces, and indica major inbred cultivars.
| Group | N | Nt | S | θ | π | Tajima’s D |
|---|---|---|---|---|---|---|
| Oryza nivara | 167 | 3 102 | 74 | 0.01323 | 0.00896 | 0.76345 |
| Indica landrace | 33 | 3 102 | 11 | 0.00946 | 0.00543 | 0.14335 |
| Indica major inbred cultivar | 193 | 3 102 | 9 | 0.00621 | 0.00354 | 0.08786 |
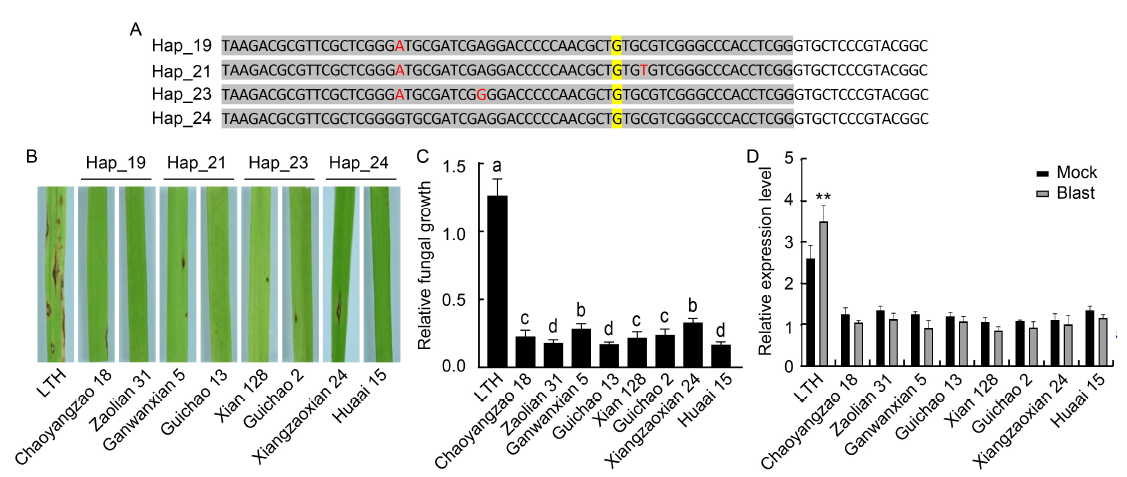
Fig. 3. Rice blast resistance analysis of four bsr-d1 resistance haplotypes. A, Nucleotide sequence alignment of four resistance haplotypes. Sites with different nucleotides among the four resistance haplotypes are marked in red font. The reported resistance site (G) is highlighted in yellow. Sites located in the promoter region are shaded in grey.B, Blast resistance of bsr-d1 resistance haplotype plants assessed by spray inoculation. Three-week-old rice plants were spray-inoculated, and photographs were taken at 7 d post-inoculation. Rice variety names are indicated below the photographs, with Lijiangxintuanheigu (LTH) as a susceptible variety control. C, Fungal growth measured as MoPot2 by qRT-PCR in the inoculated leaves, normalized to OsUbq DNA. Lowercase letters above the bars indicate multiple significant differences detected by two-way analysis of variance. Data are Mean ± SD (n = 3). D, qRT-PCR analyses performed on resistant haplotype rice accessions at 48 h post-inoculation with or without (mock) Magnaporthe oryzae. Total RNA was extracted from leaf blades of three-week-old rice plants. Ubiquitin 5 (Ubq5) was used as an internal control. Data are Mean ± SD (n = 3). P-values were determined by Student’s t-test (**, P < 0.002). Detailed information of rice cultivars is given in Table S7.
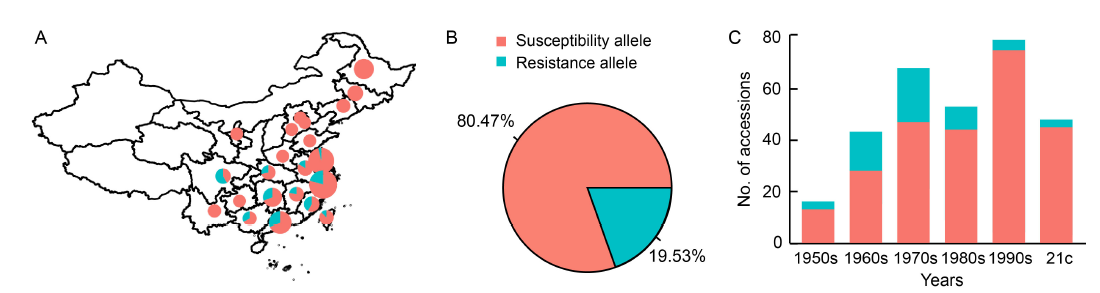
Fig. 4. Application of bsr-d1 resistance alleles in China. A, Geographic distribution of bsr-d1 resistance and susceptibility alleles among 384 major inbred cultivars in China. The size of each circle is proportional to the number of samples in each province. B, Frequency distribution of bsr-d1 resistance and susceptibility alleles in major inbred cultivars. C, Distribution of bsr-d1 resistance and susceptibility alleles in major inbred cultivars across different years. 21c, 21st century.
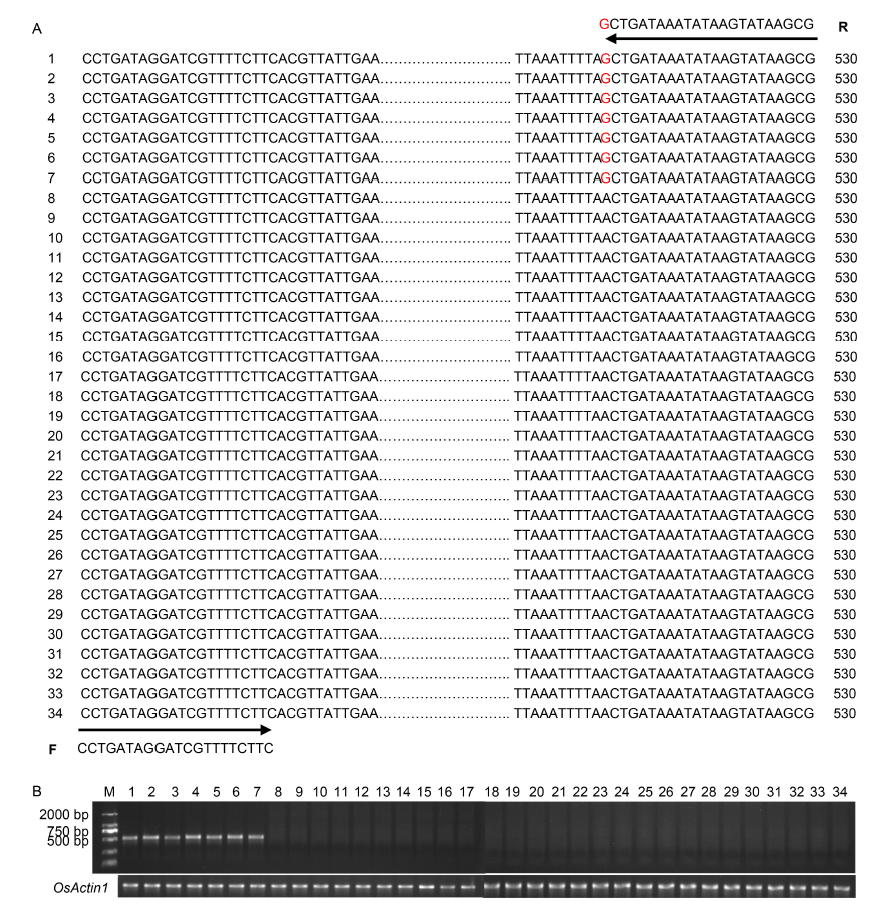
Fig. 5. Identification of bsr-d1 resistance alleles by molecular markers. A, Primer set (Bsr-d1-F/R) designed to target the single nucleotide polymorphism (SNP) site distinguishing resistance and susceptibility alleles, with an expected PCR-amplified product of 530 bp. The number shown at the left of each sequence is the rice variety code. The nucleotide polymorphic site of rice cultivars 1-7, tightly associated with rice blast resistance, is guanine (G), highlighted in red font. Varieties 8‒34 with adenine (A) are susceptible varieties. Detailed information of rice varieties is given in Table S8.B, PCR-based validation of the primer set in 34 rice varieties (30 PCR cycles). Rice varieties 1-34 are referenced in Table S8. OsActin1 (LOC_Os03g50885) was used as a control to ensure that the extracted rice genome is of good quality. M, Marker.
| [1] | Chen H L, Li C R, Liu L P, Zhao J Y, Cheng X Z, Jiang G H, Zhai W X. 2016. The Fd-GOGAT1 mutant gene lc7 confers resistance to Xanthomonas oryzae pv. Oryzae in rice. Sci Rep, 6: 26411. |
| [2] | Clement M, Posada D, Crandall K A. 2000. TCS: A computer program to estimate gene genealogies. Mol Ecol, 9(10): 1657-1659. |
| [3] | Danecek P, Auton A, Abecasis G, Albers C A, Banks E, DePristo M A, Handsaker R E, Lunter G, Marth G T, Sherry S T, McVean G, Durbin R,1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics, 27(15): 2156-2158. |
| [4] | Das A, Soubam D, Singh P K, Thakur S, Singh N K, Sharma T R. 2012. A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct Integr Genomics, 12(2): 215-228. |
| [5] | Dean R, van Kan J A L, Pretorius Z A, Hammond-Kosack K E, Di Pietro A, Spanu P D, Rudd J J, Dickman M, Kahmann R, Ellis J, Foster G D. 2012. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol, 13(4): 414-430. |
| [6] | Dean R A, Talbot N J, Ebbole D J, Farman M L, Mitchell T K, Orbach M J, Thon M, Kulkarni R, Xu J R, Pan H Q, Read N D, Lee Y H, Carbone I, Brown D, Oh Y Y, Donofrio N, Jeong J S, Soanes D M, Djonovic S, Kolomiets E, Rehmeyer C, Li W X, Harding M, Kim S, Lebrun M H, Bohnert H, Coughlan S, Butler J, Calvo S, Ma L J, Nicol R, Purcell S, Nusbaum C, Galagan J E, Birren B W. 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature, 434: 980-986. |
| [7] | Deng Y W, Zhai K R, Xie Z, Yang D Y, Zhu X D, Liu J Z, Wang X, Qin P, Yang Y Z, Zhang G M, Li Q, Zhang J F, Wu S Q, Milazzo J, Mao B Z, Wang E T, Xie H A, Tharreau D, He Z H. 2017. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science, 355: 962-965. |
| [8] | DePristo M A, Banks E, Poplin R, Garimella K V, Maguire J R, Hartl C, Philippakis A A, del Angel G, Rivas M A, Hanna M, McKenna A, Fennell T J, Kernytsky A M, Sivachenko A Y, Cibulskis K, Gabriel S B, Altshuler D, Daly M J. 2011. A framework for variation discovery and genotyping using next- generation DNA sequencing data. Nat Genet, 43(5): 491-498. |
| [9] | Fisher M C, Henk D A, Briggs C J, Brownstein J S, Madoff L C, McCraw S L, Gurr S J. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature, 484: 186-194. |
| [10] | Hong Y B, Liu Q E, Cao Y R, Zhang Y, Chen D B, Lou X Y, Cheng S H, Cao L Y. 2019. The OsMPK15 negatively regulates Magnaporthe oryza and Xoo disease resistance via SA and JA signaling pathway in rice. Front Plant Sci, 10: 752. |
| [11] | Hu S L. 2014. Analysis of the dynamics and fluctuations in the prevalence of rice blast disease in China. Liaoning Econ, (12): 90-91. (in Chinese) |
| [12] | Innan H, Kim Y. 2004. Pattern of polymorphism after strong artificial selection in a domestication event. Proc Natl Acad Sci USA, 101(29): 10667-10672. |
| [13] | Jing C Y, Zhang F M, Wang X H, Wang M X, Zhou L, Cai Z, Han J D, Geng M F, Yu W H, Jiao Z H, Huang L, Liu R, Zheng X M, Meng Q L, Ren N N, Zhang H X, Du Y S, Wang X, Qiang C G, Zou X H, Gaut B S, Ge S. 2023. Multiple domestications of Asian rice. Nat Plants, 9(8): 1221-1235. |
| [14] | Kou Y J, Wang S P. 2010. Broad-spectrum and durability: Understanding of quantitative disease resistance. Curr Opin Plant Biol, 13(2): 181-185. |
| [15] | Leigh J W, Bryant D. 2015. Popart: Full-feature software for haplotype network construction. Methods Ecol Evol, 6(9): 1110-1116. |
| [16] | Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res, 49(W1): W293-W296. |
| [17] | Li J B, Sun Y D, Liu H, Wang Y Y, Jia Y L, Xu M H. 2015. Natural variation of rice blast resistance gene Pi-d2. Genet Mol Res, 14(1): 1235-1249. |
| [18] | Li W, Zhong S H, Li G J, Li Q, Mao B Z, Deng Y W, Zhang H J, Zeng L J, Song F M, He Z H. 2011. Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defence. Cell Res, 21(5): 835-848. |
| [19] | Li W T, Liu Y, Wang J, He M, Zhou X G, Yang C, Yuan C, Wang J C, Chern M, Yin J J, Chen W L, Ma B T, Wang Y P, Qin P, Li S G, Ronald P, Chen X W. 2016. The durably resistant rice cultivar Digu activates defence gene expression before the full maturation of Magnaporthe oryzae appressorium. Mol Plant Pathol, 17(3): 354-368. |
| [20] | Li W T, Zhu Z W, Chern M, Yin J J, Yang C, Ran L, Cheng M P, He M, Wang K, Wang J, Zhou X G, Zhu X B, Chen Z X, Wang J C, Zhao W, Ma B T, Qin P, Chen W L, Wang Y P, Liu J L, Wang W M, Wu X J, Li P, Wang J R, Zhu L H, Li S G, Chen X W. 2017. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell, 170(1): 114-126.e15. |
| [21] | Li W T, Wang K, Chern M, Liu Y C, Zhu Z W, Liu J, Zhu X B, Yin J J, Ran L, Xiong J, He K W, Xu L T, He M, Wang J, Liu J L, Bi Y, Qing H, Li M W, Hu K, Song L, Wang L, Qi T, Hou Q Q, Chen W L, Li Y, Wang W M, Chen X W. 2020. Sclerenchyma cell thickening through enhanced lignification induced by OsMYB30 prevents fungal penetration of rice leaves. New Phytol, 226(6): 1850-1863. |
| [22] | Liao Y X, Bai Q, Xu P Z, Wu T K, Guo D M, Peng Y B, Zhang H Y, Deng X S, Chen X Q, Luo M, Ali A, Wang W M, Wu X J. 2018. Mutation in rice Abscisic Acid2 results in cell death, enhanced disease-resistance, altered seed dormancy and development. Front Plant Sci, 9: 405. |
| [23] | Liu Z Q, Zhu Y J, Shi H B, Qiu J H, Ding X H, Kou Y J. 2021. Recent progress in rice broad-spectrum disease resistance. Int J Mol Sci, 22(21): 11658. |
| [24] | Ma J, Lei C L, Xu X T, Hao K, Wang J L, Cheng Z J, Ma X D, Ma J, Zhou K N, Zhang X, Guo X P, Wu F Q, Lin Q B, Wang C M, Zhai H Q, Wang H Y, Wan J M. 2015. Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol Plant Microbe Interact, 28(5): 558-568. |
| [25] | Park C H, Chen S B, Shirsekar G, Zhou B, Khang C H, Songkumarn P, Afzal A J, Ning Y S, Wang R Y, Bellizzi M, Valent B, Wang G L. 2012. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell, 24(11): 4748-4762. |
| [26] | Price M N, Dehal P S, Arkin A P. 2010. FastTree 2: Approximately maximum-likelihood trees for large alignments. PLoS One, 5(3): e9490. |
| [27] | Rozas J, Sánchez-DelBarrio J C, Messeguer X, Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, 19(18): 2496-2497. |
| [28] | Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123(3): 585-595. |
| [29] | Xie Z, Yan B X, Shou J Y, Tang J, Wang X, Zhai K R, Liu J Y, Li Q, Luo M Z, Deng Y W, He Z H. 2019. A nucleotide-binding site-leucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice. Philos Trans R Soc Lond B Biol Sci, 374: 20180308. |
| [30] | Ye J H, Zhang M C, Yuan X P, Hu D X, Zhang Y Y, Xu S L, Li Z, Li R S, Liu J R, Sun Y F, Wang S, Feng Y, Xu Q, Yang Y L, Wei X H. 2022. Genomic insight into genetic changes and shaping of major inbred rice cultivars in China. New Phytol, 236(6): 2311-2326. |
| [31] | Zhao H J, Wang X Y, Jia Y L, Minkenberg B, Wheatley M, Fan J B, Jia M H, Famoso A, Edwards J D, Wamishe Y, Valent B, Wang G L, Yang Y N. 2018. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat Commun, 9(1): 2039. |
| [32] | Zhao Q, Feng Q, Lu H Y, Li Y, Wang A H, Tian Q L, Zhan Q L, Lu Y Q, Zhang L, Huang T, Wang Y C, Fan D L, Zhao Y, Wang Z Q, Zhou C C, Chen J Y, Zhu C R, Li W J, Weng Q J, Xu Q, Wang Z X, Wei X H, Han B, Huang X H. 2018. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet, 50(2): 278-284. |
| [33] | Zhu Z W, Yin J J, Chern M, Zhu X B, Yang C, He K W, Liu Y C, He M, Wang J, Song L, Wang L, Wei Y J, Wang J C, Liu J L, Qing H, Bi Y, Li M W, Hu K, Qi T, Hou Q Q, Chen X W, Li W T. 2020. New insights into bsr-d1-mediated broad-spectrum resistance to rice blast. Mol Plant Pathol, 21(7): 951-960. |
| [34] | Zhu Z W, Xiong J, Shi H, Liu Y C, Yin J J, He K W, Zhou T Y, Xu L T, Zhu X B, Lu X, Tang Y Y, Song L, Hou Q Q, Xiong Q, Wang L, Ye D H, Qi T, Zou L J, Li G B, Sun C H, Wu Z Y, Li P L, Liu J L, Bi Y, Yang Y H, Jiang C X, Fan J, Gong G S, He M, Wang J, Chen X W, Li W T. 2023. Magnaporthe oryzae effector MoSPAB1 directly activates rice Bsr-d1 expression to facilitate pathogenesis. Nat Commun, 14(1): 8399. |
| [1] | Suhas Gorakh Karkute, Vishesh Kumar, Mohd Tasleem, Dwijesh Chandra Mishra, Krishna Kumar Chaturvedi, Anil Rai, Amitha Mithra Sevanthi, Kishor Gaikwad, Tilak Raj Sharma, Amolkumar U. Solanke. Genome-Wide Analysis of von Willebrand Factor A Gene Family in Rice for Its Role in Imparting Biotic Stress Resistance with Emphasis on Rice Blast Disease [J]. Rice Science, 2022, 29(4): 375-384. |
| [2] | Singh Sarao Preetinder, Sanmallappa Bentur Jagadaish. Antixenosis and Tolerance of Rice Genotypes Against Brown Planthopper [J]. Rice Science, 2016, 23(2): 96-103. |
| [3] | GAO Qing-song, ZHANG Dan, XU Liang, XU Chen-wu. Systematic Identification of Rice ABC1 Gene Family and Its Response to Abiotic Stress [J]. RICE SCIENCE, 2011, 18(3): 167-177. |
| [4] | O. I. OLADELE, T. WAKATSUKI . Sawah Rice Eco-technology and Actualization of Green Revolution in West Africa: Experiences from Nigeria and Ghana [J]. RICE SCIENCE, 2010, 17(3): 168-172 . |
| [5] | ZHANG Qi. Genetics and Improvement of Bacterial Blight Resistance of Hybrid Rice in China [J]. RICE SCIENCE, 2009, 16(2): 83-92 . |
| [6] | GUO Xin-yi, XU Guo-hua, ZHANG Yang, HU Wei-min, FAN Long-jiang. Small-Scale Duplications Play a Significant Role in Rice Genome Evolution [J]. RICE SCIENCE, 2005, 12(3): 173-178 . |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||