#These authors contributed equally to this work
The transport of phosphate between cytoplasm and subcellular compartments is critical for plant metabolic regulation. We conducted bioinformatic analysis, heterologous expression in yeast, gene expression pattern and subcellular localization analysis to characterize the possible functions of OsPHT4 gene family in rice. Together with the PHT4 genes from higher plants, OsPHT4s can be classified into six distinct groups. OsPHT4;1-OsPHT4;4 are targeted to chloroplasts, and OsPHT4;6-1 and OsPHT4;6-2 are located to Golgi apparatus. OsPHT4 proteins can mediate inorganic phosphate (Pi) transport in yeast. In addition, dynamic transcriptional changes determined by qRT-PCR revealed different expression profiles of OsPHT4 genes in response to phosphate starvation, salicylic acid, abscisic acid and salt stress treatments. These results suggested that OsPHT4 proteins are involved in Pi distribution between the cytoplasm and chloroplast or Golgi apparatus and also involved in stress responses.
Phosphorus is an essential macronutrient required for plant growth and development, which involves in many key metabolic pathways in plants, such as photosynthesis, carbon partitioning, energy metabolism, signal transduction and enzymatic regulation (Lin et al, 2009). In higher plants, phosphorus is acquired from the soil in the form of inorganic phosphate (Pi: PO43-, HPO42-and H2PO4-) through a H+-dependent co-transport process (Raghothama, 1999). PHOSPHATE TRANS- PORTER1 (PHT1)-type proteins are defined as the major contributors to the Pi uptake system (Ayadi et al, 2015). After being absorbed through root cells, phosphate is transported within the different plant tissues and organelles by different phosphate transporters.
The phosphate transporter genes have been reported in various plant species. Based on their evolutionary relationships, the phosphate transporters can be divided into four groups (PHT1‒PHT4). The PHT1 gene family encodes Pi transporters that mediate Pi acquisition across the plasma membrane of root cells (Zhang Z L et al, 2014). The Arabidopsis AtPHT1 gene family contains nine members (AtPHT1; 1-AtPHT1; 9), among which AtPHT1; 1 to AtPHT1; 4 are the main contributors to Pi uptake from the soil to roots (Mudge et al, 2002; Ayadi et al, 2015). AtPHT1; 5 plays a significant role in Pi translocation between source to sink organs (Nagarajan et al, 2011). AtPHT1; 8 and AtPHT1; 9 are involved in Pi uptake and translocation from roots to shoots, especially under Pi-deficient conditions (Remy et al, 2012; Lapis-Gaza et al, 2014).
The rice OsPHT1 gene family has 13 members. Except for OsPHT1; 5, OsPHT1; 7 and OsPHT1; 12, all the other OsPHT1 genes have been functionally characterized (Srivastava et al, 2018; Chang et al, 2019). OsPHT1; 1 and OsPHT1; 8 have a higher affinity for Pi and are involved in Pi uptake and translocation in rice under Pi-replete conditions (Jia et al, 2011; Sun et al, 2012). Moreover, OsPHT1; 8 is also implicated in Pi remobilization from source to sink organs and Pi allocation between the embryo and endosperm of seeds (Li et al, 2015), and involved in auxin response in rice (Jia et al, 2017). Expression levels of both low-affinity OsPHT1; 2 and high-affinity OsPHT1; 6 are significantly increased under Pi deficient conditions in roots and shoots (Ai et al, 2009). OsPHT1; 6 plays a broad role in Pi uptake, translocation and internal transport throughout the plant, whereas OsPHT1; 2 mediates only Pi transport from roots to shoots (Zhang F et al, 2014). OsPHT1; 3 is strongly upregulated under Pi-deficient conditions in rice roots and shoots and mediates Pi uptake, translocation and remobilization under extremely low phosphate levels (Chang et al, 2019). OsPHT1; 4 facilitates Pi acquisition and mobilization, and also plays an important role in development of rice embryos (Ye et al, 2015; Zhang et al, 2015). OsPHT1; 9 and OsPHT1; 10 are high-affinity Pi transporters and redundantly involved in Pi uptake in rice (Wang et al, 2014). OsPHT1; 11 and OsPHT1; 13 play a central role during the development of arbuscular mycorrhizal symbioses, and OsPHT1; 11 is involved in symbiotic Pi absorption (Yang et al, 2012).
The PHT2 gene family contains one member in Arabidopsis and rice, respectively. AtPHT2; 1 mediates Pi transport into chloroplasts (Versaw and Harrison, 2002). OsPHT2; 1, a putative low affinity phosphate transporter gene, is involved in the Pi accumulation and translocation in rice (Shi et al, 2013). The Arabidopsis AtPHT3 gene family contains three members, which are localized in mitochondrial inner membranes and transport Pi into mitochondria (Zhu et al, 2012). In rice, the PHT3 gene family contains six members, however, their exact function and subcellular location are still unknown (Liu et al, 2011). The PHT4 gene family has six members in Arabidopsis. AtPHT4; 1 is located in chloroplast thylakoid membranes and acts as a Na+-dependent Pi transporter that exports Pi from thylakoid lumens to chloroplasts (Pavon et al, 2008; Karlsson et al, 2015). AtPHT4; 2 is localized in plastids and mediates Pi export from root plastids (Guo et al, 2008a; Irigoyen et al, 2011; Mukherjee et al, 2015). AtPHT4; 3 is predicted to locate in plastids and has Pi transport activity (Guo et al, 2008a). AtPHT4; 4 and AtPHT4; 5 are localized in leaf chloroplast envelope membrane as a Na+-dependent Pi transporter, and AtPHT4; 4 also exhibits significant ascorbate uptake activity (Guo et al, 2008b; Miyaji et al, 2015). AtPHT4; 6 is a Golgi-located phosphate transporter that is involved in salt tolerance and disease resistance (Cubero et al, 2009; Hassler et al, 2016).
During the past few years, several transporter genes of PHT4 family in rice have been identified (Guo et al, 2008b). However, the Pi transport function and subcellular location of them still remain elusive. In this study, we conducted a comprehensive analysis to identify members of the phosphate transporter PHT4 gene family in rice and investigated their expression patterns in various tissues, their expression profiles under Pi-starvation conditions, as well as subcellular localization of their proteins and Pi transport activity in yeast. Moreover, we examined the transcriptional profiles of the OsPHT4 genes under abiotic stress conditions. This study will lay a foundation for better understanding of the functional diversity of the OsPHT4 gene family in rice.
The protein sequences of PHT4s from different plant species were retrieved from PLAZA (http://bioinformatics. psb.ugent.be/plaza/) and the sequences were aligned with the Geneious software. Phylogenetic analysis was conducted with CIPRES (www.phylo.org) using maximum-likelihood phylogenetic analysis with 1000 bootstrap replicates.
A wild type rice variety Shishoubaimao (SSBM) was used for all physiological experiments. Hydroponic experiments were conducted as described previously (Deng et al, 2014). The Pi deprived (-P) nutrient solution had a similar composition as described in Deng et al (2014), except that NaH2PO4 was replaced by equivalent concentrations of NaCl. All the plants were grown in a greenhouse with a 12 h day (30 º C) / 12 h night (22 º C) photoperiod, nearly 200 μ mol/(m2∙ s) photon density, and approximately 60% humidity. The nutrient solution was constantly aerated and refreshed every other week.
After two weeks, seedlings were used for abiotic stress treatment. Abscisic acid (ABA, 100 mmol/L), NaCl (100 mmol/L) and salicylic acid (SA, 500 μ mol/L) were added into the cultural media. Under these stress treatments, shoot and root samples were harvested at 0, 2, 4, 8 and 24 h after treatments, frozen in liquid nitrogen, and kept at -80 º C for expression analysis.
Total RNA was extracted using the EastepTM Super Total RNA Extraction Kit (Promega, USA). For phosphate starvation treatment, the wild type plants were grown under Pi-replete conditions (200 mmol/L) for 2 weeks and then transferred to Pi starvation conditions (0 mmol/L) for 5 and 15 d. Total RNA was collected from shoots. For SA (500 μ mol/L), ABA (100 mmol/L) and NaCl (100 mmol/L) treatments, 14-day-old seedlings grown under normal conditions were exposed to different chemical treatments for 0, 4, 6, 8 and 24 h. Total RNA was collected from roots. RNA quality and quantity were determined using a Nanodrop 2000 spectrophotometer (Thermo Scientific, USA). According to the manufacturer’ s protocol, the first-strand cDNA was synthesized from 2 μ g of total RNA using the Reverse Transcription System Kit A3500 (Promega, USA) in a 20 μ L reaction volume. It was then stored at -20 º C until further use.
qRT-PCR was performed in 384-well plates on a Real-Time PCR machine (Roche, Switzerland). The 10 μ L reaction mixture contained 1 μ L cDNA diluted 5-fold, 0.5 μ mol/L of each gene-specific primer (Supplemental Table 1), and 5 μ L master mix (Light Cycler 480 SYBR Green I Master, Roche, Switzerland). The cycling condition was as follows: pre-denaturation at 95 º C for 5 min; then 40 cycles at 95 º C for 10 s (denaturation), 60 º C for 10 s (annealing), and 72 º C for 10 s (extension), followed by a melting curve analysis to confirm the correct amplification of target gene fragments and the lack of primer dimers. The rice actin gene was used as a reference and the transcript levels of genes were calculated according to the 2-Δ Δ Ct method (Pfaffl, 2001). All qRT-PCR experiments were performed with three biological replicates and three technical replicates.
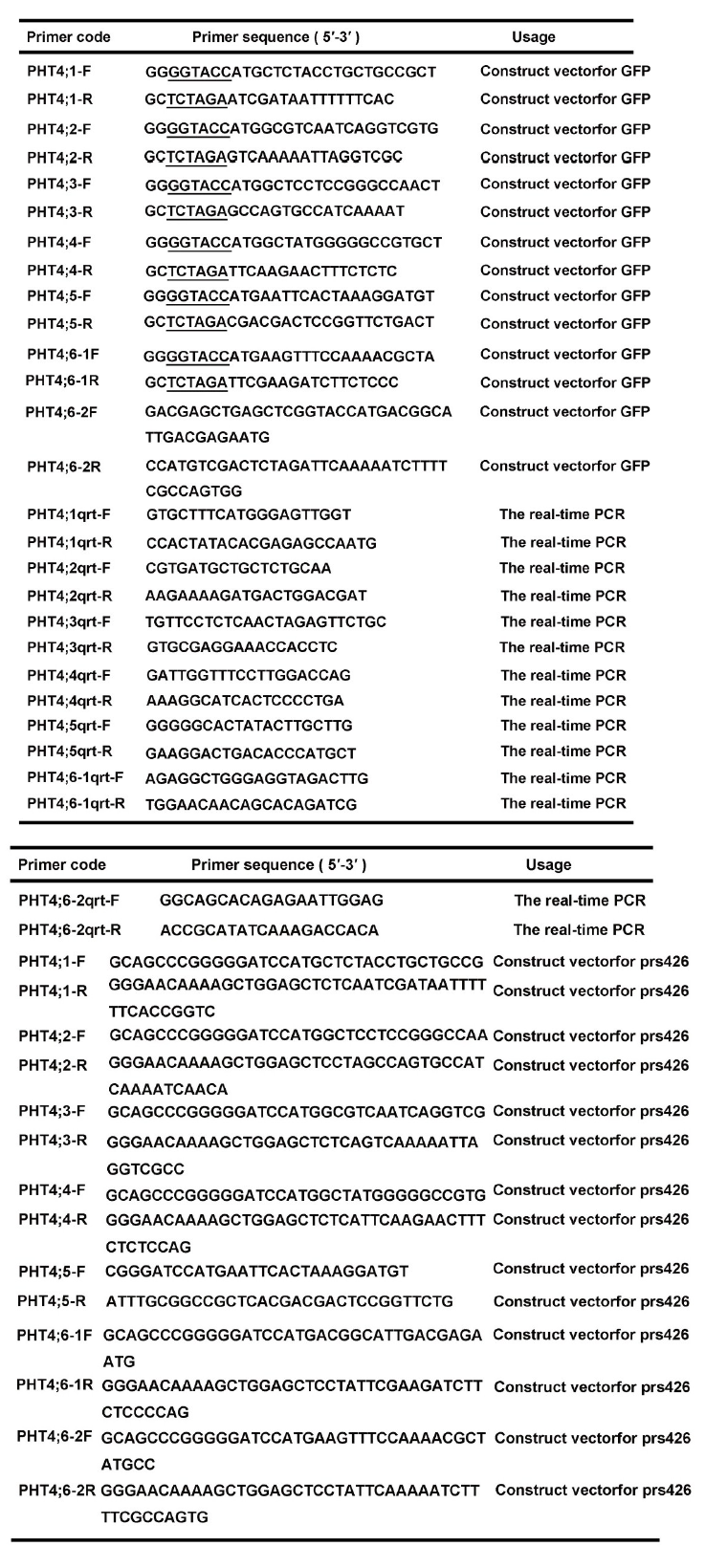
| Supplemental Table 1. Gene-specific primers used in this study. |
For subcellular localization analysis, the open reading frames (ORFs) of OsPHT4; 1, OsPHT4; 2, OsPHT4; 3, OsPHT4; 4, OsPHT4; 5, OsPHT4; 6-1andOsPHT4; 6-2 were amplified and introduced respectively into a pCAMBIA1300 vector containing 35S promoter and green fluorescent protein (GFP) fragment. The GFP was fused to the C-terminals of these genes. Primer sequences are listed in Supplemental Table 1.
The GFP fusion plasmids were transformed into protoplasts by the polyethylene glycol (PEG)-mediated transformation. Protoplasts preparation and transfection followed previously described procedures (Wu et al, 2009; Zhang et al, 2011). A Golgi localized marker was used for colocalization analysis (Nelson et al, 2007). Confocal microscopy images were taken under a laser scanning confocal microscope (Leica TCS SP5, Leica, Germany). Excitation and emission wavelengths Ex488/ Em500-550, Ex516/Em600-650 and Ex587/Em602-652 were used for GFP, autofluorescence of chlorophyll and red fluorescent protein (RFP), respectively.
To generate vectors for yeast complementation, the coding regions of PHO84, OsPHT4; 1, OsPHT4; 2, OsPHT4; 3, OsPHT4; 4, OsPHT4; 5, OsPHT4; 6-1 and OsPHT4; 6-2were introduced into the BamHI and NotI sites on a PRS426-ADH1 vector. Primers for vector constructs are listed in Supplemental Table 1. These constructs and the empty vector were transformed into a yeast mutant strain YP100 (Δ pho84Δ pho87Δ pho89 Δ pho90Δ pho91Δ git1) that is defective in Pi transport. Transformants were selected on uracil-defi cient medium and grown in synthetic complete (SC-uracil) yeast medium containing 2% galactose, 0.67% yeast nitrogen base without amino acids, 0.2% appropriate amino acids, and 2% agar at pH 5.5 and incubated at 30 º C for 3‒4 d. One colony was selected from each transformation strain and grown in the liquid SC-uracil medium. The mid-exponential growth phase cells were harvested and washed three times with sterile water, and resuspended to OD600 = 1. Equal volumes of 10-fold serial dilutions were spotted to yeast nitrogen base without amino acids (YNB) medium (without phosphate) with 2% glucose, 2% agar, appropriate amino acids and different Pi concentrations. Plates were incubated at 30 º C for 4 d.
The promoter sequences of OsPHT4 family genes were obtained from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu). We conducted the cis-element analysis using the PLACE (http://www. dna.affrc.go.jp/PLACE/). The P1BS for phosphorus deficiency response and ABA responsive element (ABRE) for ABA response were further presented.
OsPHT4; 1 (LOC_Os01g17240), OsPHT4; 2 (LOC_ Os05g37820), OsPHT4; 3 (LOC_Os01g63290), OsPHT4; 4 (LOC_Os09g39680), OsPHT4; 5(LOC_ Os09g38410), OsPHT4; 6-1 (LOC_Os11g08370), OsPHT4; 6-2 (LOC_ Os12g07970), SQD2 (AT5G01220), OsbZIP23 (LOC_ Os02g52780) and WRKY45(LOC_Os05g25770) were used.
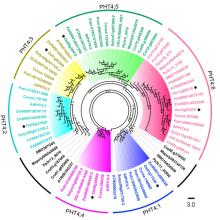
We identified and retrieved PHT4 sequences from 12 different land plants and 1 chlorophyta genomes, and examined the phylogenetic relationships between the OsPHT4 family proteins and the other PHT4 proteins. Based on genome database searching and phylogenetic analysis, seven putative PHT4 members in rice were identified and be classified into six groups (PHT4; 1‒ PHT4; 6) (Fig. 1). The PHT4 gene family from early land plants and chlorophyta formed two distinct clades, one together with PHT4; 2 and PHT4; 3 clades and the other together with PHT4; 6 clade. The results indicated that there are two orthologs of AtPHT4; 6 in rice, and we renamed them as OsPHT4; 6-1 and OsPHT4; 6-2. Together, these results clearly demonstrated that OsPHT4s share high similarities with the corresponding PHT4 proteins from Arabidopsis and other higher plants, which might have been risen from the genome duplication in the angiosperm after the separation from the early land plants.
To determine the expression patterns of OsPHT4 genes, we first checked their spatial-temporal expression profiles in RiceXPro (https://ricexpro.dna.affrc.go.jp/ GGEP/). It showed that OsPHT4; 1, OsPHT4; 4 and OsPHT4; 5were strongly expressed in leaves. OsPHT4; 2 displayed higher expression in roots and stems. OsPHT4; 3, OsPHT4; 6-1and OsPHT4; 6-2 were constitutively expressed through all tissues (Supplemental Fig. 1).
 | Supplemental Fig. 1. Transcript profiles of PHT4 family members in spatial temporal expression data in RiceXPro (https://ricexpro.dna.affrc.go.jp/GGEP/). |
We also examined the expression patterns of OsPHT4 family genes by qRT-PCR at the vegetative and reproductive growth stages of rice plants. As shown in Fig. 2, transcript levels of the OsPHT4 genes were differential in various organs of rice. At the vegetative stage, very weak expression levels of OsPHT4; 1, OsPHT4; 3, OsPHT4; 4 and OsPHT4; 5were detected in roots and the expression levels of OsPHT4; 1, OsPHT4; 3, OsPHT4; 4 and OsPHT4; 5 in shoots were higher than those in roots. Expressions of OsPHT4; 2, OsPHT4; 6-1 and OsPHT4; 6-2 in shoots were relatively high. Different from those at the vegetative stage, all OsPHT4 genes showed higher transcript levels in leaves at the flowering and grain filling stages, while very low transcript amounts were found in roots except for OsPHT4; 2 and OsPHT4; 6. In contrast, OsPHT4; 2 and OsPHT4; 6-1 exhibited higher transcript levels than OsPHT4; 1, OsPHT4; 3and OsPHT4; 4 in spikelets at the grain filling stage. In addition, the expression levels of OsPHT4; 1, OsPHT4; 3 and OsPHT4; 4 in new leaves were higher than those in old leaves.
Together, it demonstrated that the expression levels of OsPHT4 family genes vary in different organs at the different developmental stages.
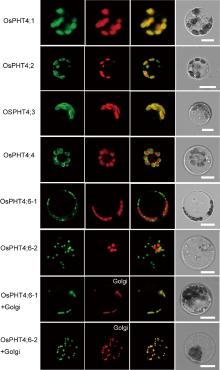
To determine the subcellular localization of the OsPHT4s, full-length complementary DNAs (cDNAs) of OsPHT4 genes were fused to GFP and transiently expressed in rice protoplasts. Confocal microscopy analysis revealed that OsPHT4; 1 was colocalized with autofluorescence of chlorophyll, which was very similar to that of the thylakoid membrane (Fig. 3). The fluorescent signals of OsPHT4; 2, OsPHT4; 3 and OsPHT4; 4 were present outside chlorophyll. The pattern of localization of these proteins was very similar with that of AtPHT4; 4, which is a chloroplast envelope membrane protein. However, OsPHT4; 6-1 and OsPHT4; 6-2 displayed punctate GFP signals that did not colocalize with the autofluorescence of chlorophyll. Given that AtPHT4; 6 is a Golgi localized protein, we introduced the GFP-OsPHT4; 6-1/GFP- OsPHT4; 6-2 construct and a Golgi marker into rice protoplasts for transient expression. Confocal microscopy analysis revealed that the signals of OsPHT4; 6-1 and OsPHT4; 6-2 were colocalized with Golgi markers, confirming the Golgi localization of OsPHT4; 6-1 and OsPHT4; 6-2 (Fig. 3). These results showed that OsPHT4; 1 was localized in chloroplast thylakoid membrane, OsPHT4; 2, OsPHT4; 3 and OsPHT4; 4 on chloroplast envelope membrane, while OsPHT4; 6-1 and OsPHT4; 6-2 on Golgi apparatus (Fig. 3). The localization of OsPHT4; 5 remained unresolved because green fluorescence was not detected in rice protoplasts. Expression of OsPHT4; 5 in rice protoplasts failed to yield detectable GFP signals, suggesting that the fusion proteins may be unstable.
These results demonstrated that OsPHT4 and AtPHT4 have similar subcellular, indicating that they may have the same function in transporting phosphate.
All the six members of Arabidopsis AtPHT4 family can complement the growth defect of PAM2 strain, a yeast Pi transport mutant defective in high-affinity Pi transporter genes PHO84 and PHO89 and facilitate Pi transport across membranes in yeast (Guo et al, 2008a). Thus, we hypothesized that the similar OsPHT4 proteins in rice might also have Pi transport functions. To test this hypothesis, we expressed OsPHT4; 1‒ OsPHT4; 6 driven by the ADH1 promoter in a yeast mutant strain YP100 that is defective in all Pi transporters and does not grow unless supplemented with galactose. It showed that the Pi transport functions of OsPHT4; 5 cannot be determined because OsPHT4; 5 was toxic to YP100 and cannot grow after transformation into yeast (Fig. 4). Transformation of the YP100 strain with the other OsPHT4 genes can all restore the growth on media containing Pi concentrations above 10 mmol/L, while transformation with empty vector did not restore the growth (Fig. 4). It indicated that the OsPHT4s can mediate Pi transport across membranes in yeast. Together, these data implied that OsPHT4; 1- OsPHT4; 6 can facilitate Pi transport across membranes in the subcellular organelles in rice.
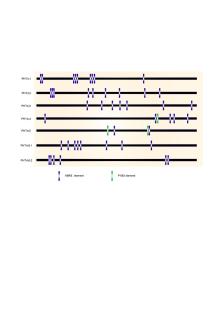
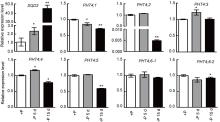
In Arabidopsis, none of the AtPHT4 genes exhibits a change in the expression level during Pi-deprived conditions for 3 d, suggesting that these genes are not responsive to Pi deprivation (Guo et al, 2008a). We also investigated whether the transcription of OsPHT4 genes was influenced by Pi supply. Since MYB transcription factors Phosphase Starvation Response can bind to the PHR1-binding sequences (P1BS) to activate the transcription of Pi starvation-induced genes and the P1BS is present in the promoter of most Pi starvation induced genes (Rubio et al, 2001), we first analyzed the P1BS in promoters of PHT4 genes using PLACE (http://www.dna.affrc.go.jp/PLACE/). It showed that most of the OsPHT4 genes had no P1BS element, except one P1BS element was detected in the promoters of OsPHT4; 4 and OsPHT4; 5 (Supplemental Fig. 2). It indicated that the expression of most of OsPHT4s might not be affected by Pi supply. Therefore, we conducted qRT-PCR to further analyze the OsPHT4 expression in rice. Consistent with the previous report (Xu et al, 2019), the expression of SQD2, encoding a sulfolipid synthase that is involved in phosphate starvation response, was induced by the phosphate starvation under the treatment. Different from SQD2, the expression levels of most of the OsPHT4 genes were only slightly affected at 5 d of phosphate starvation (Fig. 5). In contrast, the expression levels of OsPHT4; 1, OsPHT4; 2, OsPHT4; 4 andOsPHT4; 5were inhibited under Pi-deprived conditions for 15 d, suggesting that these genes might be involved in late responses to phosphate deprivation in rice shoots.
To determine whether the expression of OsPHT4 genes can be affected by abiotic stress, we firstly analyzed the ABRE cis-elements in the promoters of OsPHT4 genes using PLACE. We found that all the OsPHT4 genes contain several ABRE elements in their promoters (Supplemental Fig. 2). Therefore, the expression levels of OsPHT4 genes were investigated under ABA stress and NaCl treatment to explore their potential roles in stress tolerance.
OsbZIP23 was used to control the efficiency of ABA treatments (Xiang et al, 2008). Similar to previous report, OsbZIP23was induced by ABA in roots. The expressions of OsPHT4; 1, OsPHT4; 2, OsPHT4; 4 andOsPHT4; 5 in roots were induced by ABA, while those of OsPHT4; 3 andOsPHT4; 6-1 were only slightly induced by ABA. However, the expression level of OsPHT4; 6-2did not increase in roots exposed to ABA treatment (Fig. 6-A).
It has been reported that OsbZIP23was also induced by NaCl treatment (Xiang et al, 2008). Consistent with it, we found that the expression of OsbZIP23 was induced by the NaCl stress under our treatments. It reached peak at 4 h of NaCl treatment. Similar to it, all the OsPHT4 genes were induced in roots at 2 to 4 h after NaCl treatment (Fig. 6-B). The transcripts of OsPHT4; 1, OsPHT4; 4 and OsPHT4; 5 were increased rapidly and peaked at 2 h after NaCl treatment, then declined gradually. Other OsPHT4 genes displayed similar expression pattern and peaked at 4 h as OsbZIP23 upon NaCl treatment (Fig. 6-B).
Since SA also involved in stress response in plants, we also determined the expression levels of the OsPHT4 genes under SA treatment. As a positive control, OsWRKY45 was induced by SA in roots, which is consistent with previous report (Ryu et al, 2006). Different from NaCl treatment, only OsPHT4; 1, OsPHT4; 2, OsPHT4; 5 and OsPHT4; 6-1 were highly induced in roots by SA treatment. The root expression levels of OsPHT4; 1 and OsPHT4; 5 peaked at 4 h after SA treatment, while those of OsPHT4; 2 and OsPHT4; 6-1 peaked at 8 h after SA treatment (Fig. 6-C).
Together, it showed that the expression of OsPHT4 genes can be affected by diverse abiotic stresses.
Phosphate transporters are required for phosphate transport and uptake in plants. A systematic analysis of the whole PHT4 gene family in rice was conducted in this study, which will provide an opportunity to find candidate genes that play roles in phosphate use efficiency and abiotic stress response. A total of seven PHT4 genes are identified in rice and show quite high similarity to their corresponding members of the PHT4 gene family in Arabidopsis (70%‒80% similarities) (Guo et al, 2008b). This implies that these PHT4 genes are evolutionarily conserved and functionally similar in rice and Arabidopsis.
The expression patterns of PHT4 genes in different tissues have been described in Arabidopsis, nevertheless, the expression patterns of OsPHT4 genes in rice reveal species-specific aspects. In Arabidopsis, for example, the expressions of AtPHT4; 1 and AtPHT4; 4were detected predominantly in leaves and flowers, and at a lower level in roots; AtPHT4; 2 was constitutively expressed in roots, but was not detected in leaves and floral tissues; AtPHT4; 3 and AtPHT4; 5 exhibited similar expression patterns mainly in the vascular bundles of leaves and cotyledons; the expressions of AtPHT4; 3 and AtPHT4; 5 differed in roots and flowers, and weak expression of AtPHT4; 3 was detected at the root tip, whereas no expression of AtPHT4; 5 was detected in roots (Guo et al, 2008a). In flowers, the expression of AtPHT4; 5 was restricted to sepals, but no expression of AtPHT4; 3 was detected in flowers; AtPHT4; 6 was found to be expressed throughout the plant (Guo et al, 2008b). In contrast, at the vegetative stage, OsPHT4; 2, OsPHT4; 6-1 and OsPHT4; 6-2 had very high transcript levels in shoots and roots, but OsPHT4; 1, OsPHT4; 3, OsPHT4; 4 and OsPHT4; 5 were mainly expressed in shoots. At the flowering stage, OsPHT4; 1, OsPHT4; 3, OsPHT4; 4 and OsPHT4; 5 showed higher transcript levels in leaves than in roots, while OsPHT4; 2, OsPHT4; 6-1 and OsPHT4; 6-2 were highly expressed in roots and leaves. In addition, OsPHT4; 1, OsPHT4; 3 and OsPHT4; 4 were mainly expressed in new leaves. OsPHT4; 1, OsPHT4; 2, OsPHT4; 4 and OsPHT4; 5 showed low expression levels in spikelets at the flowering stage. At the grain filling stage, most of the OsPHT4 genes were expressed in leaves, roots and spikelets, except that OsPHT4; 1 was almost not expressed in roots and spikelets, and OsPHT4; 2 had a relatively low expression level in leaves (Fig. 2). According to the qRT-PCR data of rice, OsPHT4 genes exhibited incongruous expression patterns in various tissues, indicating that different OsPHT4 genes may have diverse functions.
Although the PHT4 genes have been studied in Arabidopsis, the physiological relevance of this family in rice is still unknown. Here, we found that like AtPHT4 proteins, the rice OsPHT4s were also located in chloroplasts or Golgi. OsPHT4; 1 was located in the chloroplast thylakoid similar to AtPHT4; 1. OsPHT4; 2, OsPHT4; 3 and OsPHT4; 4 were localized on the chloroplast envelope membranes, which suggested that the encoded proteins may have overlapping or redundant functions in chloroplasts and inactivation of multiple genes may be required to determine their physiological roles. However, OsPHT4; 2 and OsPHT4; 3 were also expressed in roots, suggesting that the proteins are also targeted to heterotrophic plastids. By contrast, OsPHT4; 6-1 and OsPHT4; 6-2 colocalized with a Golgi marker, which are consistent with AtPHT4; 6. The PHT4 gene family from early land plants and chlorophyta formed two distinct clades, one together with PHT4; 2 and PHT4; 3 clade and the other together with PHT4; 6 clade, which suggested that the divergence in subcellular localization and the function of PHT4 genes may have been occurred in the ancestor of chlorophyta.
To determine whether OsPHT4 genes have the same function of transport Pi as AtPHT4 genes, we expressed OsPHT4 proteins in a yeast Pi uptake defective mutant. Complementation of the growth defect of this mutant under different Pi conditions demonstrated that all the OsPHT4 proteins are indeed capable of mediating Pi transport, except for OsPHT4; 5 (Fig. 4). OsPHT4 proteins exhibited low affinity for Pi transport in yeast cells, which is consistent with the relatively high Pi concentrations between the cytosol and other subcellular compartments in plant cells (Mimura, 1999).
In the present study, the transcriptional patterns of OsPHT4 genes in response to abiotic stress have demonstrated that most OsPHT4 genes were responsive to phosphate deficiency, salt, SA and ABA stresses. The transcript levels of a few OsPHT4 genes, such as OsPHT4; 1, OsPHT4; 2, OsPHT4; 4 and OsPHT4; 5were inhibited in shoots of rice upon 15 d of phosphate deprivation (Fig. 5). Our experimental data showed quite similar trends with the transcriptome results from Secco et al (2013), where the expression levels of the OsPHT4 genes in shoots were not changed during short time phosphate starvation but slightly inhibited under long term of Pi-deprived conditions. This suggested that these OsPHT4 genes might respond to phosphate deficiency later and participate in phosphate efflux/import phosphate from chloroplasts in shoots of rice. Our results showed that most OsPHT4 genes are significantly or slightly up-regulated by salt, SA and ABA stresses (Fig. 6). Together, these data suggested that the phosphate transporters OsPHT4 gene family is very important for salt tolerance, disease resistance and ABA signal transduction. In accordance with our results, the Atpht4; 6mutant was hypersensitive to NaCl stress, as shown by root growth inhibition and swelling in root elongation area, implying that AtPHT4; 6 is also involved in salt stress response (Cubero et al, 2009). A loss-of-function mutation in the AtPHT4; 6 gene enhanced disease resistance to Pseudomonas syringae infection and accumulated high levels of SA, suggesting that AtPHT4; 6is important for defense responses (Hassler et al, 2012). Over-expression of AtPHT4; 1gene in wild-type conferred enhanced susceptibility to P. syringae infection, suggesting that AtPHT4; 1 is also critical for basal defense (Wang et al, 2011). Most of the OsPHT4 genes were remarkably induced by ABA in roots, indicating that OsPHT4 genes might be participated in abiotic stress responses. Therefore, it is interesting to explore the roles of OsPHT4 genes in ABA signal by using corresponding rice mutants and overexpressed genetic materials.
Pi transporters are located in cell membranes and catalyze the transport of Pi from one compartment to another. In plants, the PHT1 gene family encodes Pi transporters that transport Pi across the plasma membrane of root cells located at the root-soil interface. The PHT2, PHT3 and PHT4 gene families encode proteins that transport Pi between the cytoplasm and different subcellular compartments. PHT2 is a low-affinity phosphate transporter located in the inner membranes of chloroplasts. It is mainly responsible for the absorption of Pi into chloroplasts. The PHT3 family is a phosphate transporter located in mitochondria, which can transport phosphate to mitochondria. Unlike other PHT families, members of PHT4 family are located in chloroplasts or Golgi bodies. OsPHT4s can confer phosphate transport activity in yeast. Therefore, we speculated that OsPHT4; 1- OsPHT4; 6 can facilitate Pi transport across membranes in chloroplasts or Golgi bodies in rice. Meanwhile, OsPHT4 genes may be participated in abiotic stress responses. However, the physiological functions of this family in rice need further investigation.
This study was financially supported by the National Natural Science Foundation of China (Grant Nos. 31801924 and 31972492).
The following materials are available in the online version of this article at http://www.sciencedirect.com/science/journal/16726308; http://www.ricescience.org.
Supplemental Table 1. Gene-specific primers used in this study.
Supplemental Fig. 1. Transcript profiles of PHT4 family members in spatial temporal expression data in RiceXPro (https://ricexpro.dna.affrc.go.jp/GGEP/).
Supplemental Fig. 2. Analysis of OsPHT4 gene family promoter sequences.
(Managing Editor: Wu Yawen)
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|