
Rice Science ›› 2015, Vol. 22 ›› Issue (6): 255-263.DOI: 10.1016/S1672-6308(14)60307-3
• • 下一篇
收稿日期:2015-04-22
接受日期:2015-06-09
出版日期:2015-06-06
发布日期:2015-09-15
. [J]. Rice Science, 2015, 22(6): 255-263.
| Gene | ID | Chromosome | Amino acid size |
|---|---|---|---|
| OsMAX1a | LOC_Os01g50530 | 1 | 412 |
| LOC_Os01g50520 | 1 | ||
| OsMAX1b | LOC_Os06g36920 | 6 | 549 |
| OsMAX1c | LOC_Os01g50590 LOC_Os01g50580 | 1 | 517 |
| OsMAX1d | LOC_Os02g12890 | 1 | 385 |
| OsMAX1e | 2 | 548 |
Table 1 OsMAX1 candidate genes in rice.
| Gene | ID | Chromosome | Amino acid size |
|---|---|---|---|
| OsMAX1a | LOC_Os01g50530 | 1 | 412 |
| LOC_Os01g50520 | 1 | ||
| OsMAX1b | LOC_Os06g36920 | 6 | 549 |
| OsMAX1c | LOC_Os01g50590 LOC_Os01g50580 | 1 | 517 |
| OsMAX1d | LOC_Os02g12890 | 1 | 385 |
| OsMAX1e | 2 | 548 |

Fig. 1. Chromosomal location and gene structure of rice OsMAX1 candidate genes.(Chr, Chromosome. Black boxes indicate exon and the connecting lines indicate intron in part B. )
| Marker | Sence primer | Anti-sence primer |
|---|---|---|
| RNAi-OsMAX1CON-F RNAi-OsMAX1CON-R | 5'-GCGTCGACGTTCTCAAGAGGCTTCGCTG-3' | 5'-CGAGCTCGTTCTCAAGAGGCTTCGCTG-3' |
| RNAi-OsMAX1a-F | 5'-CCCAAGCTTCAAGGTAGGGGAATTTGGTCT-3' | 5'-CGGAATTCCAAGGTAGGGGAATTTGGTCT-3' |
| RNAi-OsMAX1a-R | 5'-GCGTCGACTCCTGAAGGAAGCGATGAGA-3' | 5'-CGAGCTCTCCTGAAGGAAGCGATGAGA-3' |
| RNAi-OsMAX1e-F | 5'-CCCAAGCTTCACCTCTGGCTCTGGGAAAT-3' | 5'-CGGAATTCCACCTCTGGCTCTGGGAAAT-3' |
| RNAi-OsMAX1e-R | 5'-GCGTCGACGTCCAGCTCGCTGTCCACCA-3' | 5'-TCCCCCGGGGTCCAGCTCGCTGTCCACCA-3' |
| 5'-CCCAAGCTTGAGACGAGGTGGAAGCGCATG-3' | 5'-CGGAATTCGAGACGAGGTGGAAGCGCATG-3' |
Table 2 Sequence of RNAi markers.
| Marker | Sence primer | Anti-sence primer |
|---|---|---|
| RNAi-OsMAX1CON-F RNAi-OsMAX1CON-R | 5'-GCGTCGACGTTCTCAAGAGGCTTCGCTG-3' | 5'-CGAGCTCGTTCTCAAGAGGCTTCGCTG-3' |
| RNAi-OsMAX1a-F | 5'-CCCAAGCTTCAAGGTAGGGGAATTTGGTCT-3' | 5'-CGGAATTCCAAGGTAGGGGAATTTGGTCT-3' |
| RNAi-OsMAX1a-R | 5'-GCGTCGACTCCTGAAGGAAGCGATGAGA-3' | 5'-CGAGCTCTCCTGAAGGAAGCGATGAGA-3' |
| RNAi-OsMAX1e-F | 5'-CCCAAGCTTCACCTCTGGCTCTGGGAAAT-3' | 5'-CGGAATTCCACCTCTGGCTCTGGGAAAT-3' |
| RNAi-OsMAX1e-R | 5'-GCGTCGACGTCCAGCTCGCTGTCCACCA-3' | 5'-TCCCCCGGGGTCCAGCTCGCTGTCCACCA-3' |
| 5'-CCCAAGCTTGAGACGAGGTGGAAGCGCATG-3' | 5'-CGGAATTCGAGACGAGGTGGAAGCGCATG-3' |

Fig. 3. Over-expression of OsMAX1a and OsMAX1e in Arabidopsis max1 mutant.(A and D, Over-expression of OsMAX1a in Arabidopsis max1; B and E, Over-expression of OsMAX1e in Arabidopsis max1; C and F, Arabidopsis max1.)
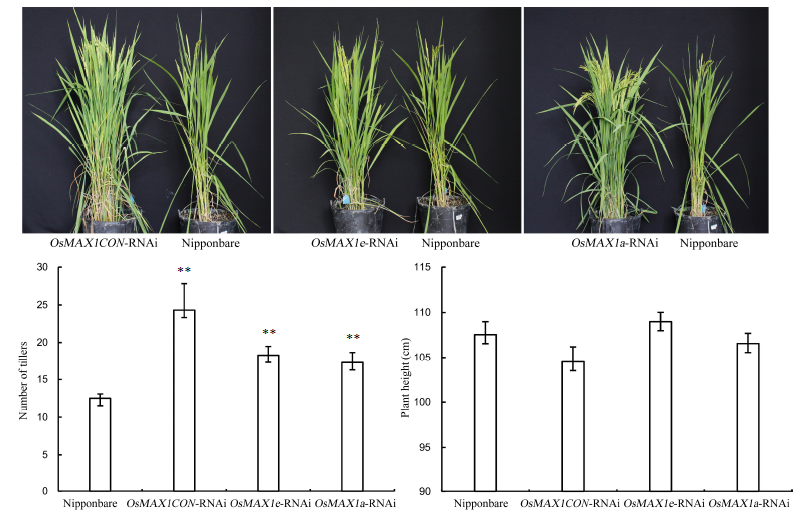
Fig. 5. Tillering numbers and plant heights of Nipponbare, OsMAX1CON, OsMAX1a and OsMAX1e interference in rice.(OsMAX1CON-RNAi, Conserved region interference of OsMAX1 gene. ** means significant difference at the 0.01 level. Bar represents the standard error.)
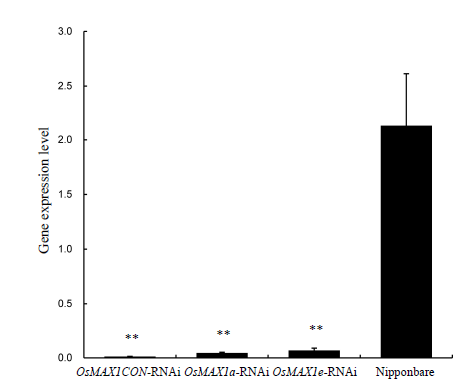
Fig. 6. Expression levels of OsMAX1CON, OsMAX1a and OsMAX1e interference offspring in rice.(** means significant difference at the 0.01 level. Bar represents the standard error.)
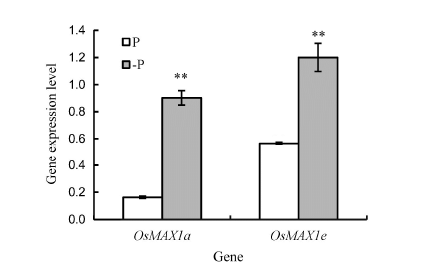
Fig. 7. Expression levels of OsMAX1a and OsMAX1e as affected by phosphate (P)-deficiency.(** means significant difference at the 0.01 level. Bar represents the standard error.)
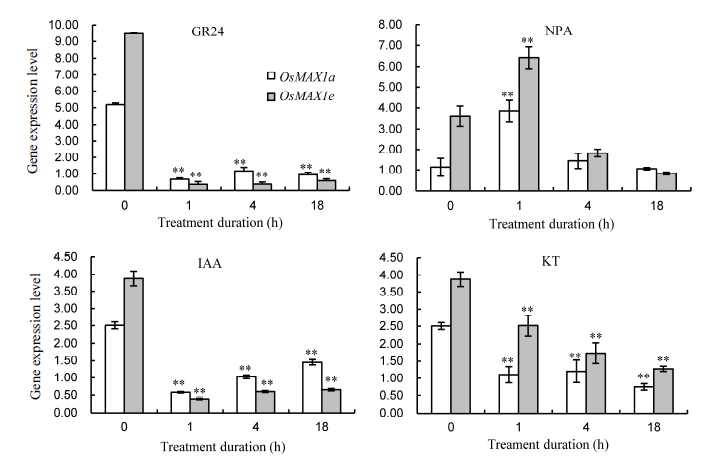
Fig. 8. Expression levels of OsMAX1a and OsMAX1e after GR24, N-1-naphthylphalamic acid (NPA), indole-3-acetic acid (IAA) and kinetin (KT) treatments.(** means significant difference at the 0.01 level. Bar represents the standard error.)
| [1] | Akiyama K, Matsuzaki K I, Hayashi H.2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi.Nature, 435: 824-827. |
| [2] | Alder A, Holdermann I, Beyer P, Al-Babili S.2008. Carotenoid oxygenases involved in plant branching catalyse a highly specific conserved apocarotenoid cleavage reaction.Biochem J, 416: 289-296. |
| [3] | Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Ai-Babili S.2012. The path from β-carotene to carlactone, a strigolactone-like plant hormone.Science, 335: 1348-1351. |
| [4] | Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaquchi S, Kyozuka J.2009. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers.Plant Cell Physiol, 50: 1416-1424. |
| [5] | Beveridge C A.2000. Long-distance signalling and a mutational analysis of branching in pea.Plant Growth Regul, 32: 193-203. |
| [6] | Booker J, Auldridge M, Wills S, Mccarty D, Klee H, Leyser O.2004. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule.Curr Biol, 14: 1232-1238. |
| [7] | Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O.2005. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone.Dev Cell, 8: 443-449. |
| [8] | Cardoso C, Ruyter-Spira C, Bouwmeester H J.2011. Strigolactones and root infestation by plant-parasitic Striga, Orobanche and Phelipanche spp.Plant Sci, 180(3): 414-420. |
| [9] | Cock J H, Yoshida S, Forno D A, Gomez K A.1976. Laboratory Manual for Physiological Studies of Rice. 3rd ed. Los Banos, the Philippines: International Rice Research Institute: 61-64. |
| [10] | Cook C E, Whichard L P, Turner B, Wall M E, Egley G H.1966. Germination of witchweed (Striga lutea Lour.): Isolation and properties of a potent stimulant.Science, 154: 1189-1190. |
| [11] | Domagalska M A, Leyser O.2011. Signal integration in the control of shoot branching.Nat Rev Mol Cell Biol, 12: 211-221. |
| [12] | Drummond R S M, Sheehan H, Simons J L, Martinez-Sanchez N M, Turner R M, Putterill J, Snowden K C.2011. The expression of petunia strigolactone pathway genes is altered as part of the endogenous developmental program.Front Plant Sci, 2: 115. |
| [13] | Dun E A, Brewer P B, Beveridge C A.2009. Strigolactones: Discovery of the elusive shoot branching hormone.Trends Plant Sci, 14: 364-372. |
| [14] | Gomez-Roldan V, Fermas S, Brewer P B, Puech-Pages V, Dun E A, Pillot J P, Letisse F, Matusova R, Danoun S, Portais J C, Bouwmeester H, Becard G, Beveridge C A, Rameau C, Rochange S F.2008. Strigolactone inhibition of shoot branching.Nature, 455: 189-194. |
| [15] | Guo S Y, Xu Y Y, Liu H H, Mao Z W, Zhang C, Ma Y, Zhang Q R, Meng Z, Chong K.2013. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14.Nat Commun, 4: 1566. |
| [16] | Hamiaux C, Drummond R S M, Janssen B J, Ledger S E, Cooney J M, Newcomb R D, Snowden K C.2012. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone.Curr Biol, 22: 2032-2036. |
| [17] | Hiei Y, Ohta S, Komari T, Kumashiro T.1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA.Plant J, 6: 271-282. |
| [18] | Huang J Q, Wei Z M, An H L, Zhu Y X.2001. Agrobacterium tumefaciens-mediated transformation of rice with the spider insecticidal gene conferring resistance to leaffolder and striped stem borer.Cell Res, 11: 149-155. |
| [19] | Jiang L, Liu X, Xiong G S, Liu H H, Chen F L, Wang L, Meng X B, Liu G F, Yu H, Yuan Y D, Yi W, Zhao L H, Ma H L, He Y Z, Wu Z S, Melcher K, Qian Q, Xu H E, Wang Y H, Li J Y.2013. DWARF53 acts as a repressor of strigolactone signalling in rice.Nature, 504: 401-405. |
| [20] | Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska M A, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C.2011. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis.Plant Physiol, 155(2): 974-987. |
| [21] | Lin H, Wang R, Qian Q, Yan M X, Meng X B, Fu Z M, Yan C Y, Jiang B, Su Z, Li J Y, Wang Y H.2009. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth.Plant Cell, 21: 1512-1525. |
| [22] | Ruyter-Spira C, Al-Babili S, Van Der Krol S, Bouwmeester H.2013. The biology of strigolactones.Trends Plant Sci, 18: 72-83. |
| [23] | Snowden K C, Simkin A J, Janssen B J, Templeton K R, Loucas H M, Simons J L, Karunairetnam S, Gleave A P, Clark D G, Klee H J.2005. The decreased apical dominance1/petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development.Plant Cell, 17: 746-759. |
| [24] | Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chtfield S, Ward S, Beveridge C, Rameau C, Levser O.2003. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea.Genes Dev, 17: 1469-1474. |
| [25] | Stirnberg P, Sande K V D, Leyser H M O.2002. MAX1 and MAX2 control shoot lateral branching in Arabidopsis.Development, 129: 1131-1141. |
| [26] | Turnbull C G, Booker J P, Leyser H M.2002. Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J, 32: 255-262. |
| [27] | Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J, Yamaguchi S.2008. Inhibition of shoot branching by new terpenoid plant hormones.Nature, 455: 195-200. |
| [28] | Wang T, Yuan S J, Yin L, Zhao J F, Wan J M, Li X Y.2013. Positional cloning and expression analysis of the gene responsible for the high-tillering dwarf phenotype in the indica rice mutant gsor23.Chin J Rice Sci, 27(1): 1-8. (in Chinese with English abstract) |
| [29] | Wang Y H, Li J Y.2011. Branching in rice.Curr Opin Plant Biol, 14: 94-99. |
| [30] | Wang Y, Sun S Y, Zhu W J, Jia K P, Yang H Q, Wang X L.2013. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching.Dev Cell, 27: 681-688. |
| [31] | Yoneyama K, Yoneyama K, Takeuchi Y, Sekimoto H.2007. Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites.Planta, 225(4): 1031-1038. |
| [32] | Zhou F, Lin Q B, Zhu L H, Ren Y L, Zhou K N, Shabek N, Wu F Q, Mao H B, Dong W, Gan L, Ma W W, Gao H, Chen J, Yang C, Wang D, Tan J J, Zhang X, Guo X P, Wang J L, Jiang L, Liu X, Chen W Q, Chu J F, Yan C Y, Ueno K, Ito S, Asami T, Cheng Z J, Wang J, Lei C L, Zhai H Q, Wu C Y, Wang H Y, Zheng N, Wan J M.2013. D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling.Nature, 504: 406-410. |
| [33] | Zou J H, Zhang S Y, Zhang W P, Li G, Chen Z X, Zhai W X, Zhao X F, Pan X B, Xie Q, Zhu L H.2006. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds.Plant J, 48(5): 687-698. |
| [34] | Xu H M, Zhang L J, Liu C.2010. Inflorescence infection of Arabidopsis by Agrobacterium.North Hort, 14: 143-146. (in Chinese with English abstract) |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||