
Rice Science ›› 2019, Vol. 26 ›› Issue (2): 129-132.DOI: 10.1016/j.rsci.2018.12.006
• • 上一篇
收稿日期:2018-10-30
接受日期:2018-12-18
出版日期:2019-03-04
发布日期:2018-12-18
. [J]. Rice Science, 2019, 26(2): 129-132.
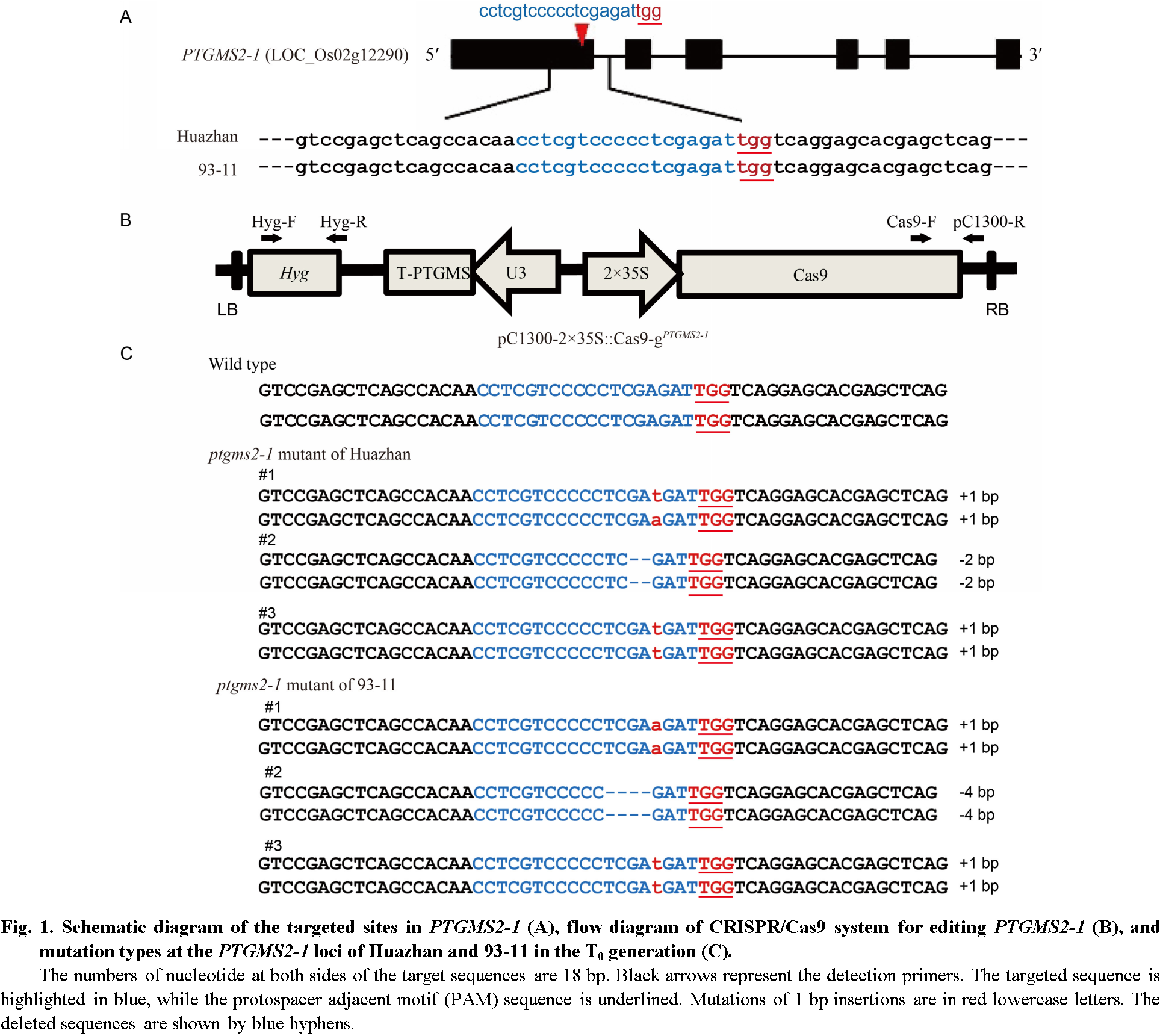
Fig. 1 Schematic diagram of the targeted sites in PTGMS2-1 (A), flow diagram of CRISPR/Cas9 system for editing PTGMS2-1 (B), and mutation types at the PTGMS2-1 loci of Huazhan and 93-11 in the T0 generation (C).The numbers of nucleotide at both sides of the target sequences are 18 bp. Black arrows represent the detection primers. The targeted sequence is highlighted in blue, while the protospacer adjacent motif (PAM) sequence is underlined. Mutations of 1 bp insertions are in red lowercase letters. The deleted sequences are shown by blue hyphens.

Fig. 2 Pollen fertility of wild type (WT) and ptgms2-1 in 93-11 and Huazhan.A–H, I2-KI staining of pollen grains of WT and ptgms2-1 in 93-11. Scale bars are 200 μm. I–P, I2-KI staining of pollen grains of WT and ptgms2-1 in Huazhan. Scale bars are 200 μm.
| Primer name | Sequence (5′-3′) | Experiment |
| PTGMS-g++ | GGCAACCTCGTCCCCCTCGAGAT | Vector construction |
| PTGMS-g-- | AAACATCTCGAGGGGGACGAGGT | |
| T3 | ATTAACCCTCACTAAAGGGA | |
| PTGMS-JC-F | GTCTCGCAGGAGTTCCTCTT | Detection of target mutations |
| PTGMS-JC-R | GAGGCCAAGATACTCTGGCT | |
| Hyg-F | AAGACCAATGCGGAGCATATAC | PCR-based genotyping of the T-DNA construct |
| Hyg-R | GTGATATCTCCACTGACGTA | |
| Cas9-F | ACCAGACACGAGACGACTAA | |
| pC1300-R | ATCGGTGCGGGCCTCTTC | |
| Actin-F | TGCTATGTACGTCGCCATCCA | |
| Actin-R | AATGAGTAACCACGCTCCGTC |
Supplemental Table 1 Primers used in this research.
| Primer name | Sequence (5′-3′) | Experiment |
| PTGMS-g++ | GGCAACCTCGTCCCCCTCGAGAT | Vector construction |
| PTGMS-g-- | AAACATCTCGAGGGGGACGAGGT | |
| T3 | ATTAACCCTCACTAAAGGGA | |
| PTGMS-JC-F | GTCTCGCAGGAGTTCCTCTT | Detection of target mutations |
| PTGMS-JC-R | GAGGCCAAGATACTCTGGCT | |
| Hyg-F | AAGACCAATGCGGAGCATATAC | PCR-based genotyping of the T-DNA construct |
| Hyg-R | GTGATATCTCCACTGACGTA | |
| Cas9-F | ACCAGACACGAGACGACTAA | |
| pC1300-R | ATCGGTGCGGGCCTCTTC | |
| Actin-F | TGCTATGTACGTCGCCATCCA | |
| Actin-R | AATGAGTAACCACGCTCCGTC |
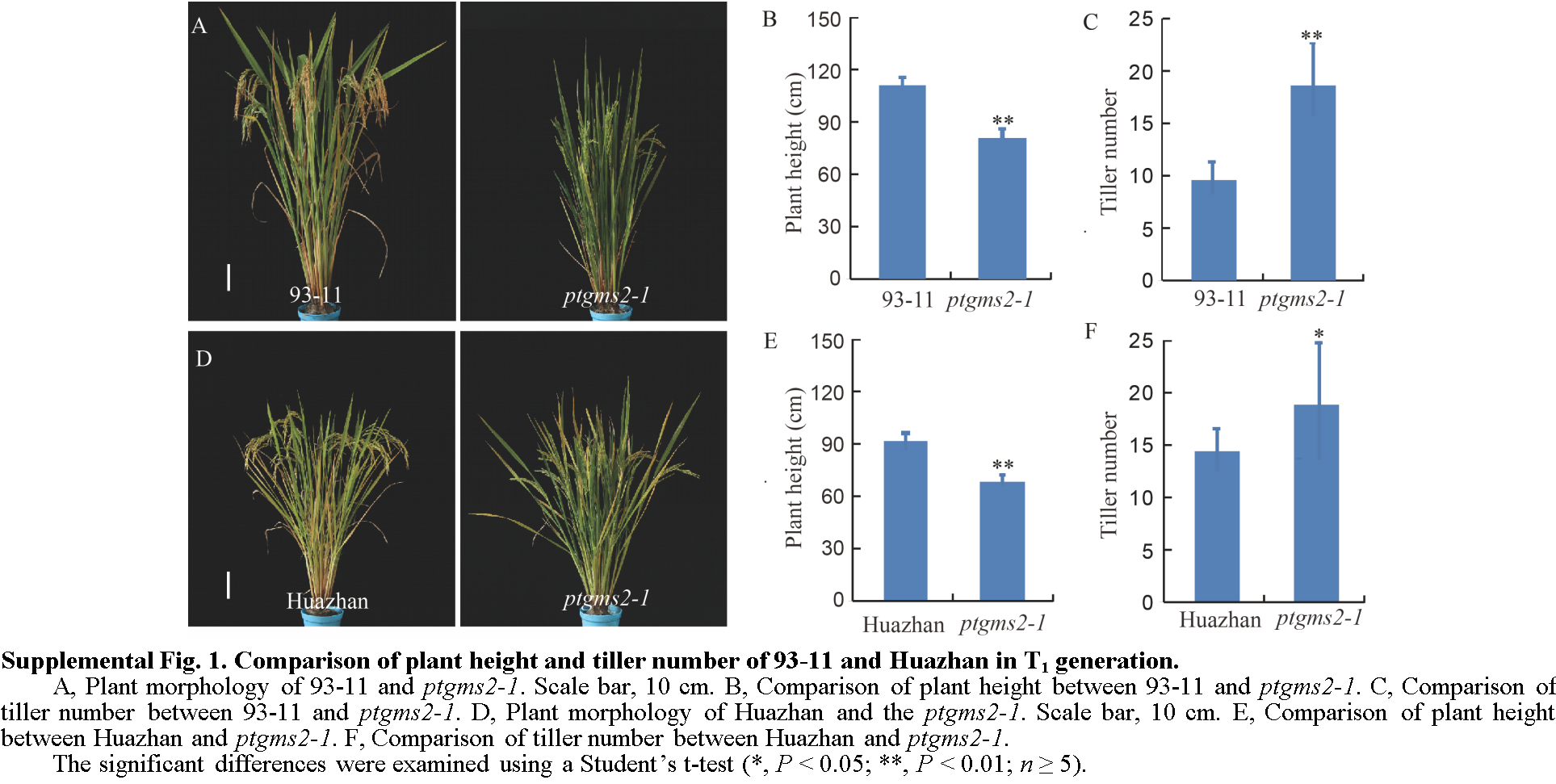
Fig. 1 Comparison of plant height and tiller number of 93-11 and Huazhan with the mutants in T1 generation.A, Plant morphology of 93-11 and ptgms2-1. Scale bar, 10 cm. B, Comparison of plant height between 93-11 and ptgms2-1. C, Comparison of tiller number between 93-11 and ptgms2-1. D, Plant morphology of Huazhan and the ptgms2-1. Scale bar, 10 cm. E, Comparison of plant height between Huazhan and ptgms2-1. F, Comparison of tiller number between Huazhan and ptgms2-1. The significant differences were examined using a Student’s t-test (*, P < 0.05; **, P < 0.01; n ≥ 5).
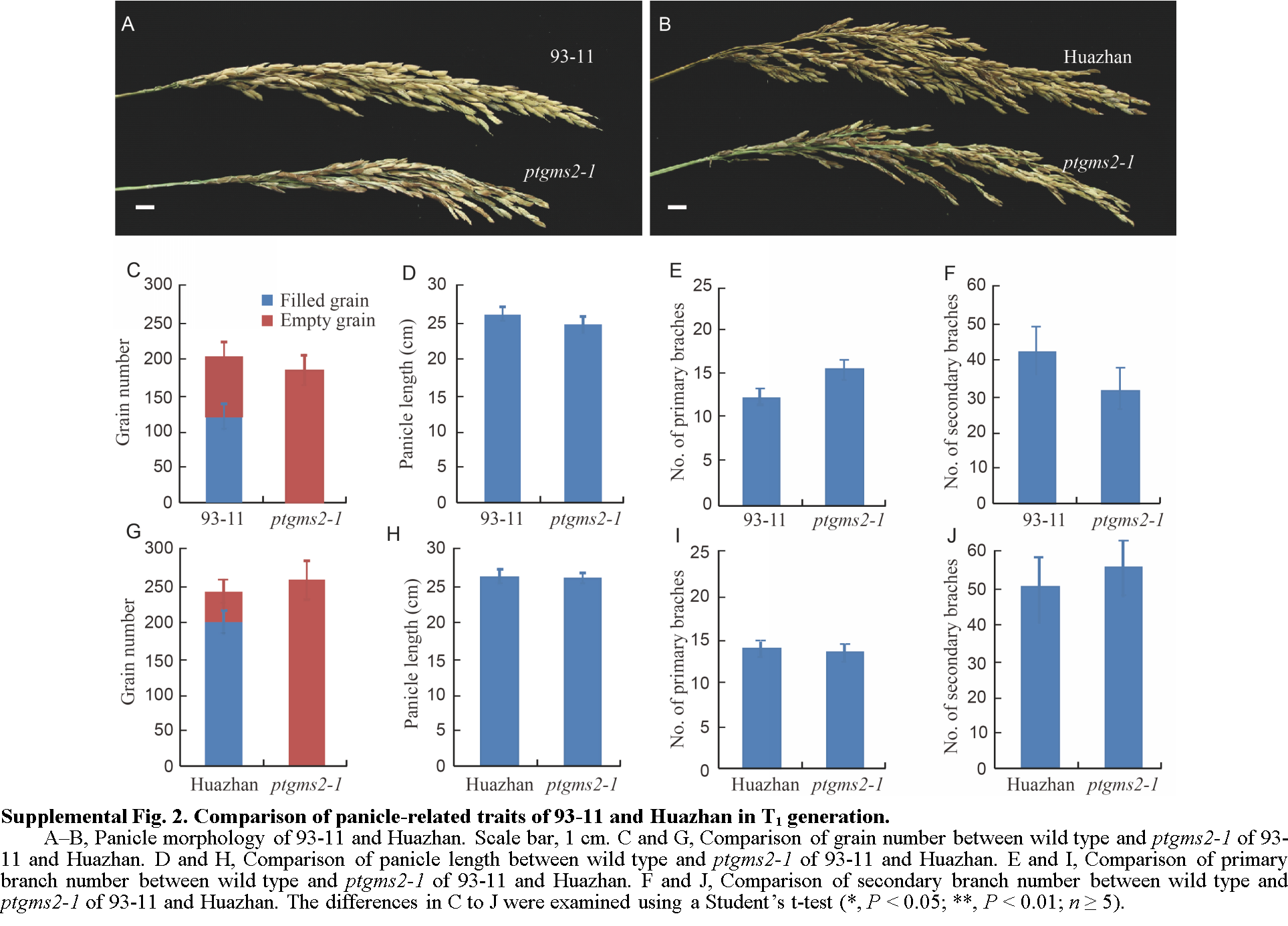
Fig. 2 Comparison of panicle-related traits of 93-11 and Huazhan with the mutants in T1 generation.A–B, Panicle morphology of 93-11 and Huazhan. Scale bar, 1 cm. C and G, Comparison of grain number between wild type and ptgms2-1 of 93-11 and Huazhan. D and H, Comparison of panicle length between wild type and ptgms2-1 of 93-11 and Huazhan. E and I, Comparison of primary branch number between wild type and ptgms2-1 of 93-11 and Huazhan. F and J, Comparison of secondary branch number between wild type and ptgms2-1 of 93-11 and Huazhan. The differences in C to J were examined using a Student’s t-test (*, P < 0.05; **, P < 0.01; n ≥ 5).
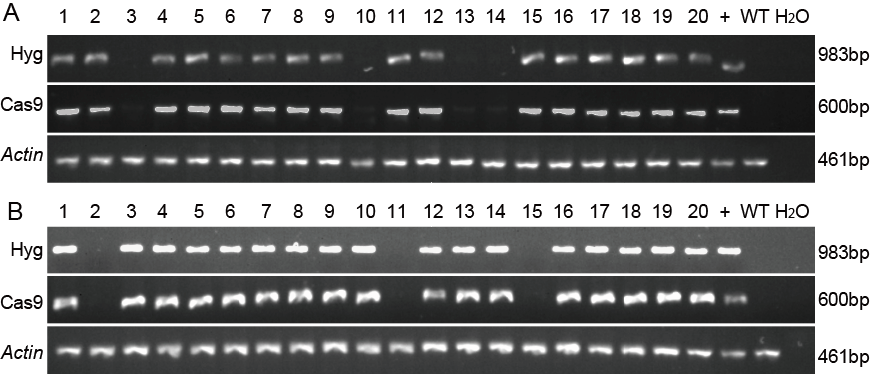
Fig. 3 PCR identification of the marker-free transgenic plants in T1 generation.A, PCR identification of the marker-free transgenic plants in T1 generation in 93-11. B, PCR identification of the marker-free transgenic plants in T1 generation in Huazhan. 1-20, T1 transgenic lines. +, Positive control of transgenic line. WT and H2O, Negative control. Actin, a gene was used as control.
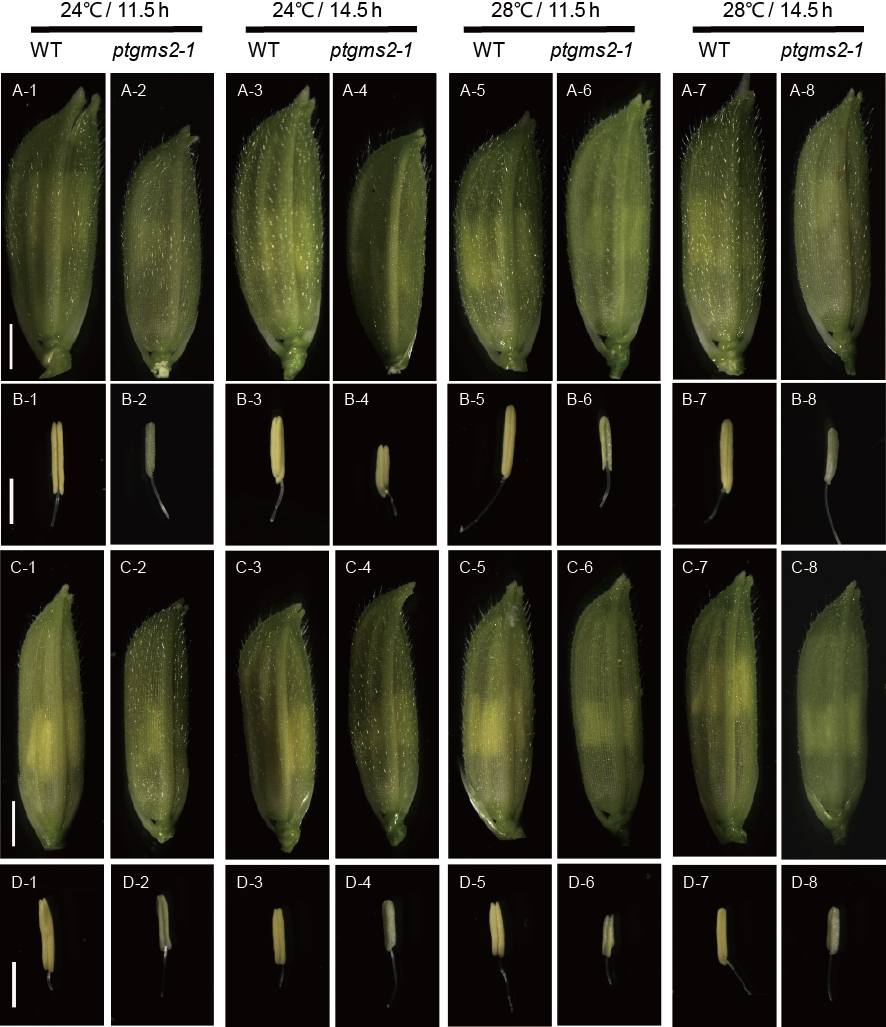
Fig. 4 Morphology of WT and ptgms2-1 in 9311 and Huazhan.A, Mature florets of 9311 and ptgms2-1 plants. Scale bars = 2 mm. B, Anther of 9311 and ptgms2-1 plants. Scale bars = 2 mm. C, Mature florets of Huazhan and ptgms2-1 plants. Scale bars = 2 mm. D, Anther of Huazhan and ptgms2-1 plants. Scale bars = 2 mm.
| [1] | Chang Z Y, Chen Z F, Wang N, Xie G, Lu J W, Yan W, Zhou J L, Tang X Y, Deng X W.2016. Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene.Proc Natl Acad Sci USA, 113(49): 14145-14150. |
| [2] | Cheng S H, Zhuang J Y, Fan Y Y, Du J H, Cao L Y.2007. Progress in research and development on hybrid rice: A super-domesticate in China.Ann Bot, 100(5): 959-966. |
| [3] | Fan Y R, Yang J Y, Mathioni S M, Yu J S, Shen J Q, Yang X F, Wang L, Zhang Q H, Cai Z X, Xu C G, Li X H, Xiao J H, Meyers B C, Zhang Q F.2016. PMS1T, producing phased small- interfering RNAs, regulates photoperiod-sensitive male sterility in rice.Proc Natl Acad Sci USA, 113(52): 15144-15149. |
| [4] | Jiang J H, Ni J L, Wu S, Wang D Z.2017. Development of permanent genic male sterile line by pyramiding thermo-sensitive genic male sterile genes and reverse temperature induced genic male sterile genes in rice (Oryza sativa L.). Chin J Rice Sci, 31(4): 371-378. (in Chinese with English abstract) |
| [5] | Peng S, Cassman K G, Virmani S S, Sheehy J, Khush G S.1999. Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential.Crop Sci, 39(6): 1552-1559. |
| [6] | Ren D Y, Hu J, Xu Q K, Cui Y J, Zhang Y, Zhou T T, Rao Y C, Xue D W, Zeng D L, Zhang G H, Gao Z Y, Zhu L, Shen L, Chen G, Guo L B, Qian Q.2018. FZP determines grain size and sterile lemma fate in rice.J Exp Bot, 69(20): 4853-4866. |
| [7] | Shen L, Wang C, Fu Y P, Wang J J, Liu Q, Zhang X M, Yan C J, Qian Q, Wang K J.2018. QTL editing confers opposing yield performance in different rice varieties.J Integr Plant Biol, 60(2): 89-93. |
| [8] | Tang L, Mao B G, Li Y K, Lv Q M, Zhang L P, Chen C Y, He H J, Wang W P, Zeng X F, Shao Y, Pan Y L, Hu Y Y, Peng Y, Fu X Q, Li H Q, Xia S T, Zhao B R.2017. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci Rep, 7(1): 14438. |
| [9] | Xu J J, Wang B H, Wu Y H, Du P N, Wang J, Wang M, Yi C D, Gu M H, Liang G H.2011. Fine mapping and candidate gene analysis of ptgms2-1, the photoperiod-thermo-sensitive genic male sterile gene in rice(Oryza sativa L.). Theor Appl Genet, 122(2): 365-372. |
| [10] | Zhang J S, Zhang H, Botella J R, Zhu J K.2018. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J Integr Plant Biol, 60(5): 369-375. |
| [11] | Zhou H, Liu Q J, Li J, Jiang D G, Zhou L Y, Wu P, Lu S, Li F, Zhu L Y, Liu Z L, Chen L T, Liu Y G, Zhuang C X.2012. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA.Cell Res, 22(4): 649-660. |
| [12] | Zhou H, Zhou M, Yang Y Z, Li J, Zhu L Y, Jiang D G, Dong J F, Liu Q J, Gu L F, Zhou L Y, Feng M J, Qin P, Hu X C, Song C L, Shi J F, Song X W, Ni E D, Wu X J, Deng Q Y, Liu Z L, Chen M S, Liu Y G, Cao X F, Zhuang C X.2014. RNase ZS1 processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice.Nat Commun, 5: 4884. |
| [13] | Zhou H, He M, Li J, Chen L, Huang Z F, Zheng S Y, Zhu L Y, Ni E D, Jiang D G, Zhao B R, Zhuang C X.2016. Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediatedTMS5 editing system. Sci Rep, 6: 37395. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||