
Rice Science ›› 2020, Vol. 27 ›› Issue (2): 86-100.DOI: 10.1016/j.rsci.2020.01.001
收稿日期:2019-06-26
接受日期:2019-09-29
出版日期:2020-03-28
发布日期:2019-11-28
. [J]. Rice Science, 2020, 27(2): 86-100.
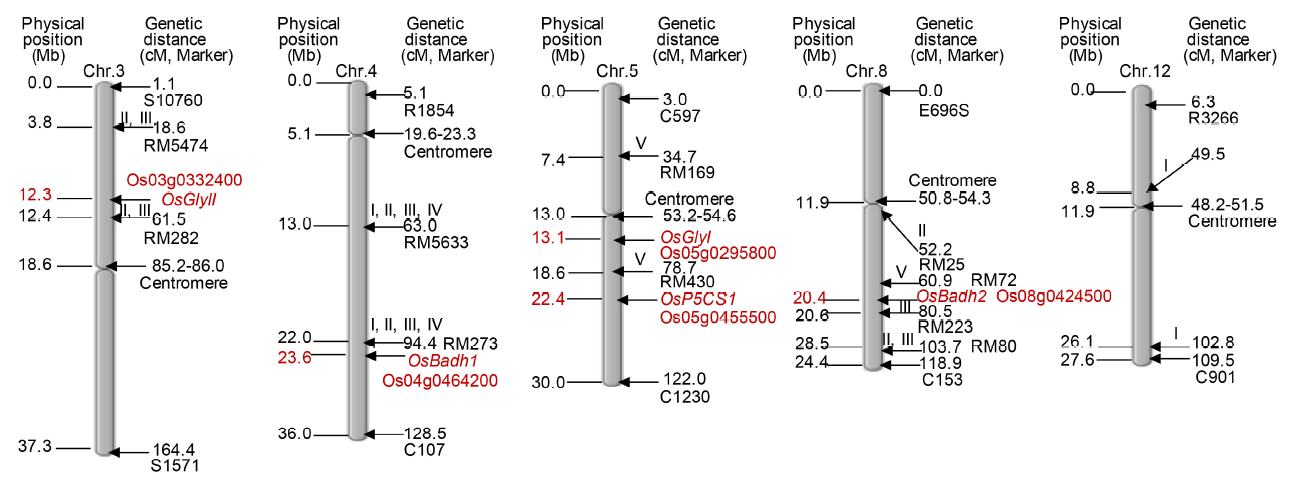
Fig. 1. Integration of previously reported QTLs for aroma with respective candidate genes in rice. I, Luriex et al (1996); II, Singh et al (2007); III, Amaranthi et al (2008); IV, Pachuri et al (2014); V, Talukder et al (2017); Chr, Chromosome; Mb, Mega base; cM, Centimorgan.The map has been constructed using the Chromosome Map Tool of Oryzabase from http://viewer.shigen.info/oryzavw/maptool/MapTool.do; http://archive.gramene.org/qtl/ and http://archive.gramene.org/markers/ (information retrieved on 20 May, 2019).
| Gene | ID | Tissue specificity (Expression) | Response to stress | Splicing | Subcellular localization | Protein interaction |
|---|---|---|---|---|---|---|
| P5CS gene | ||||||
| OsP5CS1 | Os05g0455500 LOC_Os05g38150 | Flower buds, milk grains | Osmoregulation, salinity, anoxia | Introns 3, 19; Termination | Chl, ER, Nu, Cyt, Mit, Pl, Extr, Vac | Ferredoxin-dependent glutamate synthase |
| OsP5CS2 | Os01g0848200 LOC_Os01g62900 | Flowers, flower buds | Osmoregulation, salinity, anoxia | All introns; Termination | Chl, Extr, Nu, Vac | Glutamate synthase |
| GLY gene | ||||||
| OsGlyI | Os05g0295800 LOC_Os05g22970 | Leaves before flowering, flower buds | Salinity, anoxia | ‒ | Chl, Cyt | ‒ |
| OsGlyII | Os03g0332400 LOC_Os03g21460 | Flower buds, flower | Salinity, anoxia | ‒ | Cyt, Chl, Nu, Extr | Glyoxalase |
| OsGlyIII | Os01g0667200 LOC_Os01g47690 | Roots and leaves before flowering | Salinity, anoxia | Introns 7, 8 | Chl, Per, Cyt, Golg | Ferredoxin-nitrite reductase |
| BADH gene | ||||||
| OsBadh1 | Os04g0464200 LOC_Os04g39020 | Roots before flowering, flowers | Salinity, anoxia, submergence | Intron 4 | Per, Chl, Cyt, Nu | Glutamate synthase |
| OsBadh2 | Os08g0424500 LOC_Os08g32870 | Flowers, flower buds | Salinity, anoxia, submergence | ‒ | Chl, Per, Cyt, Nu | Glutamate synthase |
Table 1 Description of some potential genes related to aroma in rice.
| Gene | ID | Tissue specificity (Expression) | Response to stress | Splicing | Subcellular localization | Protein interaction |
|---|---|---|---|---|---|---|
| P5CS gene | ||||||
| OsP5CS1 | Os05g0455500 LOC_Os05g38150 | Flower buds, milk grains | Osmoregulation, salinity, anoxia | Introns 3, 19; Termination | Chl, ER, Nu, Cyt, Mit, Pl, Extr, Vac | Ferredoxin-dependent glutamate synthase |
| OsP5CS2 | Os01g0848200 LOC_Os01g62900 | Flowers, flower buds | Osmoregulation, salinity, anoxia | All introns; Termination | Chl, Extr, Nu, Vac | Glutamate synthase |
| GLY gene | ||||||
| OsGlyI | Os05g0295800 LOC_Os05g22970 | Leaves before flowering, flower buds | Salinity, anoxia | ‒ | Chl, Cyt | ‒ |
| OsGlyII | Os03g0332400 LOC_Os03g21460 | Flower buds, flower | Salinity, anoxia | ‒ | Cyt, Chl, Nu, Extr | Glyoxalase |
| OsGlyIII | Os01g0667200 LOC_Os01g47690 | Roots and leaves before flowering | Salinity, anoxia | Introns 7, 8 | Chl, Per, Cyt, Golg | Ferredoxin-nitrite reductase |
| BADH gene | ||||||
| OsBadh1 | Os04g0464200 LOC_Os04g39020 | Roots before flowering, flowers | Salinity, anoxia, submergence | Intron 4 | Per, Chl, Cyt, Nu | Glutamate synthase |
| OsBadh2 | Os08g0424500 LOC_Os08g32870 | Flowers, flower buds | Salinity, anoxia, submergence | ‒ | Chl, Per, Cyt, Nu | Glutamate synthase |
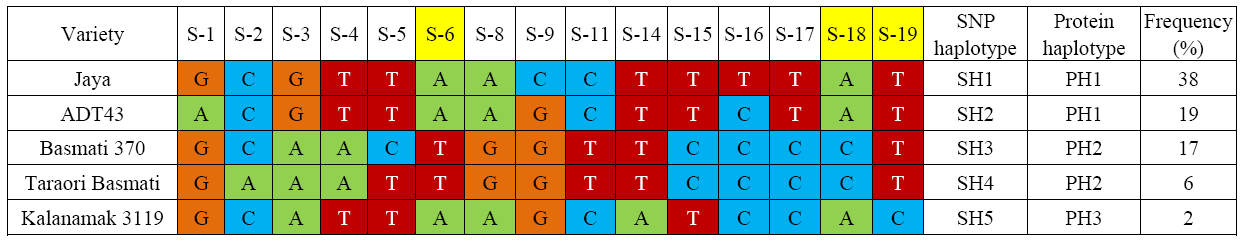
Fig. 2. Haplotypes of OsBadh1 gene in rice based on 15 single nucleotide polymorphisms (SNPs) with no missing data genotyped using Sequenom MassARRAY system. Protein haplotypes are based on three exonic SNPs (S6, S18 and S19). It was redrawn from Singh et al (2010).
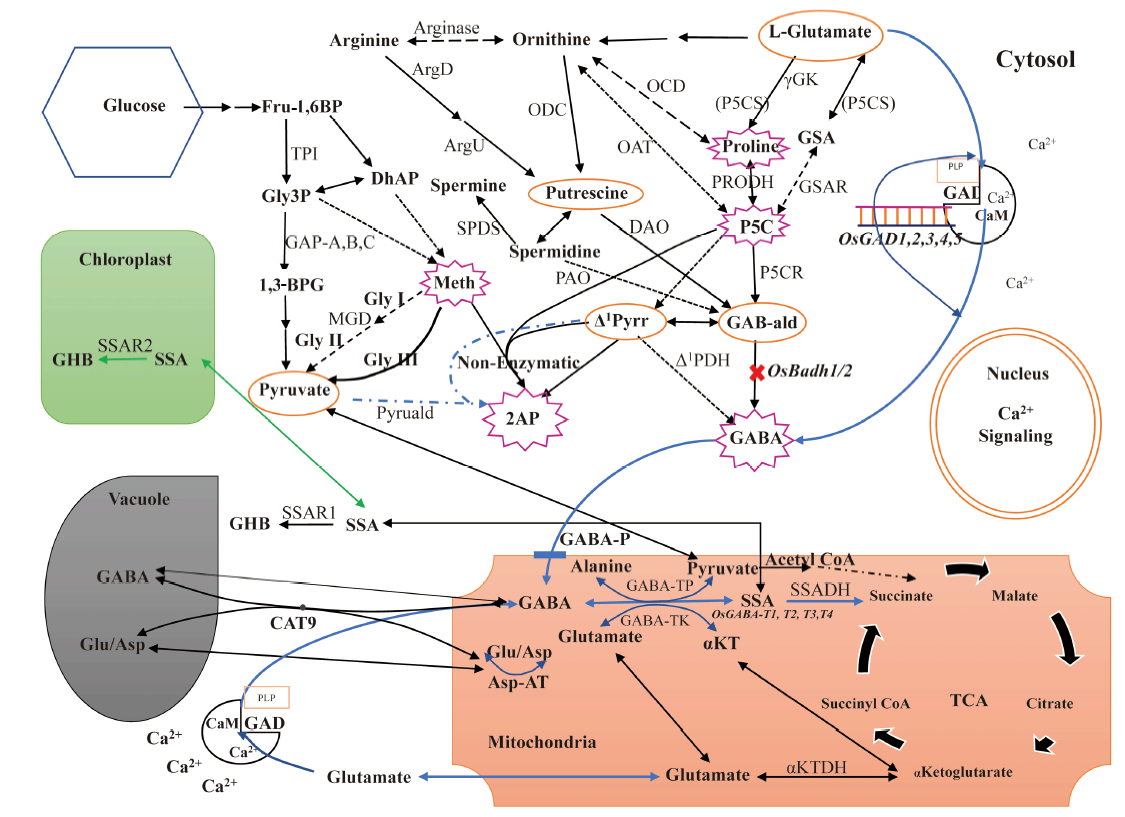
Fig. 3. Comprehensive 2-acetyl-1-phrroline (2AP) pathway related to other pathways in aromatic rice plant cells. 1,3-BPG, 1,3-biphosphoglycerate; ArgD, Arginine decarboxylase; ArgU, Argamatine ureodehydrolase; Asp, Aspartic acid; Asp-AT, Aspartate aminotransferase; CaM, Calmodulin; CAT9, Cationic amino acid transporter 9; DAO, Diamine oxidase; DhAP, Dihydroxyacetone phosphate; Fru-1,6BP, Fructose-1,6-biphosphate; GABA, γ-aminobutyric acid; GAB-ald, γ-aminobutyraldehyde; GABA-P, γ-aminobutyric acid permease; GABA-TK, α-ketoglutarate dependent GABA transaminase; GABA-TP, Pyruvate dependent GABA transaminase; GAD, Glutamate decarboxylase; GAP-A,B,C, Glyceraldehyde-3-phosphate dehydrogenase homologues (A,B,C); GHB, Gamma hydroxybutyrate; Glu, Glutamate; Gly3P, Glyceraldehyde-3-phosphate; GSA, Gamma glutamyl semialdehyde; GSAR, Gamma glutamyl semialdehyde reductase; Meth, Methylglyoxal; MGD, Methylglyoxal dehydrogenase; OAT, Ornithine aminotransferase; OCD, Ornithine cyclodeaminase; ODC, Ornithine decarboxylase; P5C, Δ1-pyrroline-5-carboxylate; P5CS, Δ1-pyrroline-5-carboxylate synthetase; P5CR, Δ1-pyrroline-5-carboxylate reductase; PAO, Polyamine oxidase; PLP, Pyridoxal 5′-phosphate; PRODH, Proline dehydrogenase; SPDS, Spermidine synthase; SSA, Succinic semialdehyde; SSADH, Succinic semialdehyde dehydrogenase; SSAR, Succinic semialdehyde reductase; TCA, Tricarboxylic acid; TPI, Triosephosphate isomerase; αKT, α-ketoglutarate; αKTDH, α-ketoglutarate dehydrogenase; γ-GK, γ-glutamyl kinase; Δ1PDH, Δ1-pyrroline dehydrogenase; Δ1-pyrr, Δ1-pyrroline.
| Treatment/Environment | Variety | 2AP concentration (µg/kg) | Reference |
|---|---|---|---|
| Shading | |||
| Control | Yuxiangyouzhan | 85.10 | |
| Treated | Yuxiangyouzhan | 175.86 | |
| Control | Nongxiang 18 | 94.84 | |
| Treated | Nongxiang 18 | 135.02 | |
| Salinity | |||
| 2006 | Aychade | 715.00 | |
| 2007 | Aychade | 1 440.00 | |
| 2006 | Fidji | 560.00 | |
| 2007 | Fidji | 1 715.00 | |
| 2006 | Giano | 301.00 | |
| 2007 | Giano | 741.00 | |
| Growing condition | |||
| Sandy soil, dry during ripening | KDML105 | 532.00 | |
| Submerged during ripening | KDML105 | 218.00 | |
| Clay soil | KDML105 | 388.00 | |
| Growth stage | |||
| Vegetative stage | AM157 | 310.00 | |
| Reproductive stage | AM157 | 400.00 | |
| Grain filling stage | AM157 | 580.00 | |
| Mature grain stage | AM157 | 662.00 | |
| Vegetative stage | BA370 | 380.00 | |
| Reproductive stage | BA370 | 420.00 | |
| Grain filling stage | BA370 | 260.00 | |
| Mature grain stage | BA370 | 451.00 |
Table 2 Variation in 2-acetyl-1-phrroline (2AP) concentration under different environmental conditions.
| Treatment/Environment | Variety | 2AP concentration (µg/kg) | Reference |
|---|---|---|---|
| Shading | |||
| Control | Yuxiangyouzhan | 85.10 | |
| Treated | Yuxiangyouzhan | 175.86 | |
| Control | Nongxiang 18 | 94.84 | |
| Treated | Nongxiang 18 | 135.02 | |
| Salinity | |||
| 2006 | Aychade | 715.00 | |
| 2007 | Aychade | 1 440.00 | |
| 2006 | Fidji | 560.00 | |
| 2007 | Fidji | 1 715.00 | |
| 2006 | Giano | 301.00 | |
| 2007 | Giano | 741.00 | |
| Growing condition | |||
| Sandy soil, dry during ripening | KDML105 | 532.00 | |
| Submerged during ripening | KDML105 | 218.00 | |
| Clay soil | KDML105 | 388.00 | |
| Growth stage | |||
| Vegetative stage | AM157 | 310.00 | |
| Reproductive stage | AM157 | 400.00 | |
| Grain filling stage | AM157 | 580.00 | |
| Mature grain stage | AM157 | 662.00 | |
| Vegetative stage | BA370 | 380.00 | |
| Reproductive stage | BA370 | 420.00 | |
| Grain filling stage | BA370 | 260.00 | |
| Mature grain stage | BA370 | 451.00 |
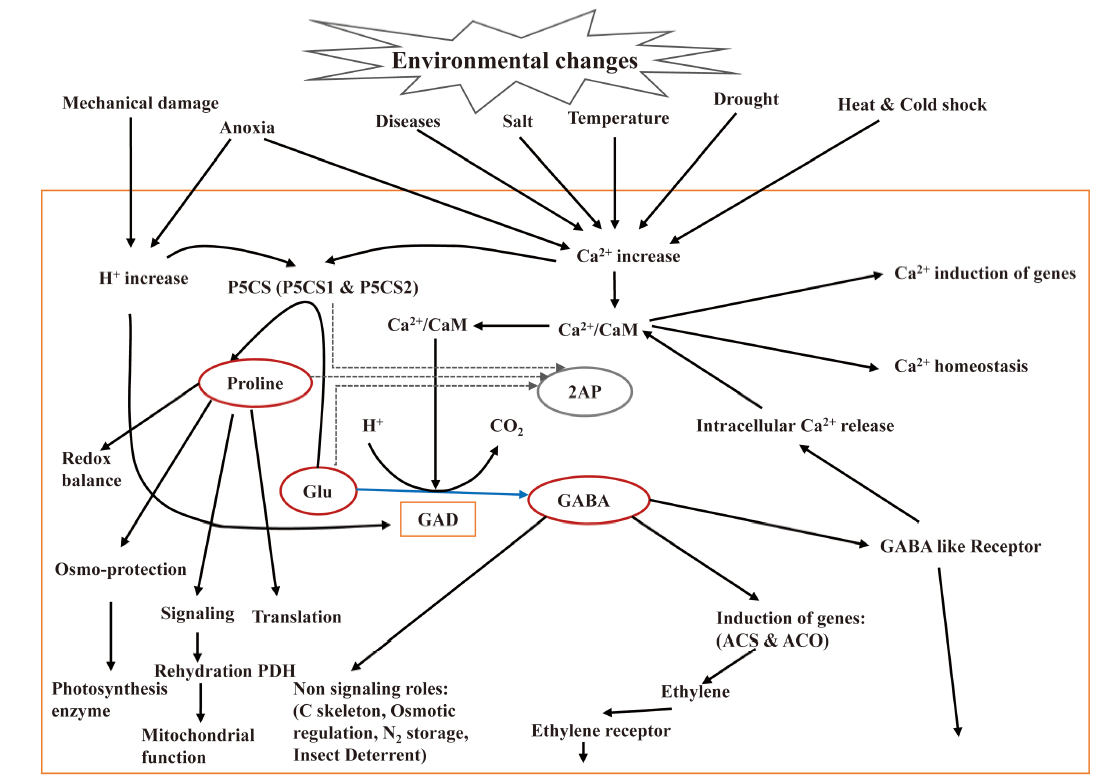
Fig. 4. Roles of GABA and proline in plant stress responses and environmental changes. 2AP, 2-acetyl-1-phrroline; ACS, 1-aminocyclopropane-1-carboxylic acid synthase; ACO, 1-aminocyclopropane-1-carboxylic acid oxidase; CaM, Calmodulin; GABA, γ-aminobutyric acid; GAD, Glutamate decarboxylase; Glu, Glutamate; P5CS, Δ1-pyrroline-5-carboxylate synthetase; PDH, Proline dehydrogenase.GABA and proline may function as a cellular barometer and transducer of environmental stress signals (Modified from Kinnersley and Turano, 2000).
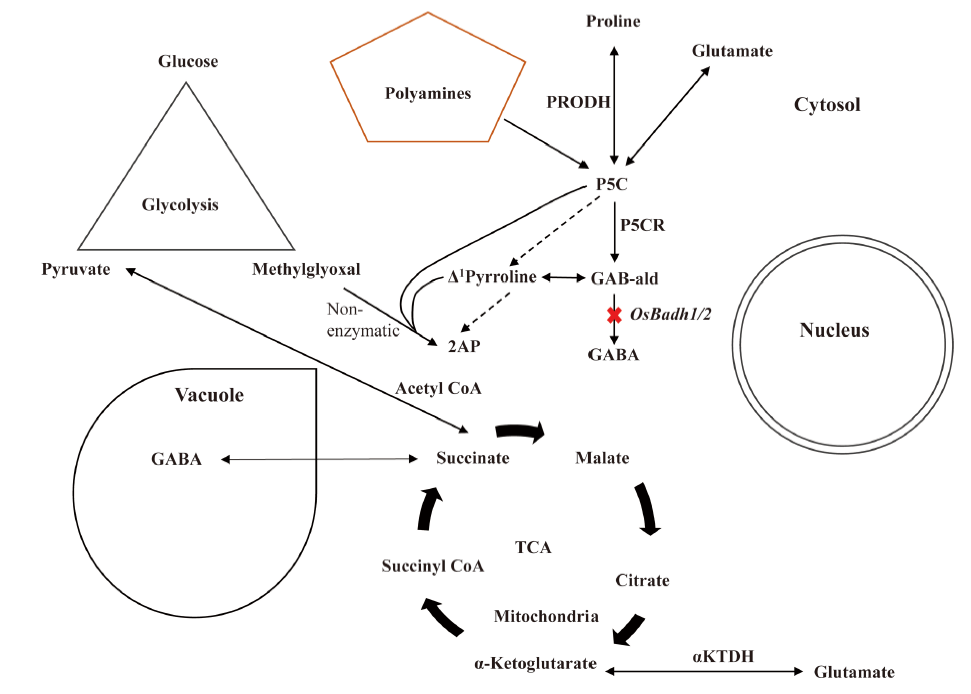
Fig. 5. Regulation of aroma related compounds during normal or optimum environmental condition.2AP, 2-acetyl-1-pyrroline; GABA, γ-aminobutyric acid; GAB-ald, Gamma aminobutyraldehyde; P5C, Δ1-pyrroline-5-carboxylate; P5CR, Δ1-pyrroline-5-carboxylate reductase; PRODH, Proline dehydrogenase; TCA, Tricarboxylic acid cycle; α-KTDH, α-ketoglutarate dehydrogenase.
| [1] | Akama K, Takaiwa F. 2007. C-terminal extension of rice glutamate decarboxylase (OsGAD2) functions as an autoinhibitory domain and overexpression of a truncated mutant results in the accumulation of extremely high levels of GABA in plant cells. J Exp Bot, 58(10): 2699-2707. |
| [2] | Amarawathi Y, Singh R, Singh A K, Singh V P, Mohapatra T, Sharma T R, Singh N K. 2008. Mapping of quantitative trait loci for Basmati quality traits in rice (Oryza sativa L.). Mol Breeding, 21(1): 49-65. |
| [3] | Bao L F, Lin G, Zhao D M, Li Y W, Zeng J, He B. 2007. Characters for indica-type and good-quality cytoplasmic male sterile line of Yixiang 1A. J Plant Genet Res, 1: 016. |
| [4] | Bartels D, Sunkar R. 2005. Drought and salt tolerance in plants. Crit Rev Plant Sci, 24(1): 23-58. |
| [5] | Baum G, Lev-Yadun S, Fridmann Y, Arazi T, Katsnelson H, Zik M, Fromm H. 1996. Calmodulin binding to glutamate decarboxylase is required for regulation of glutamate and GABA metabolism and normal development in plants. EMBO J, 15(12): 2988-2996. |
| [6] | Birla D S, Malik K, Sainger M, Chaudhary D, Jaiwal R, Jaiwal P K. 2017. Progress and challenges in improving the nutritional quality of rice (Oryza sativa L.). Crit Rev Food Sci Nutr, 57(11): 2455-2481. |
| [7] | Bourgis F, Guyot R, Gherbi H, Tailliez E, Amabile I, Salse J, Lorieux M, Delseny M, Ghesquiere A. 2008. Characterization of the major fragance gene from an aromatic japonica rice and analysis of its diversity in Asian cultivated rice. Theor Appl Genet, 117(3): 353-368. |
| [8] | Bradbury L M T, Fitzgerald T L, Henry R J, Jin Q S, Waters D L E. 2005a. The gene for fragrance in rice. Plant Biotechnol J, 3(3): 363-370. |
| [9] | Bradbury L M T, Henry R J, Jin Q S, Reinke R F, Waters D L E. 2005b. A perfect marker for fragrance genotyping in rice. Mol Breeding, 16(4): 279-283. |
| [10] | Bradbury L M T, Gillies S A, Brushett D J, Waters D L E, Henry R J. 2008. Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol Biol, 68(4-5): 439-449. |
| [11] | Bradbury L M T. 2009. Identification of the gene responsible for fragrance in rice and characterisation of the enzyme transcribed from this gene and its homologs. Lismore, NSW Australia: Southern Cross University: 1-91. |
| [12] | Buttery R G. 1982. 2-Acetyl-1-pyrroline: An important aroma component of cooked rice. Chem Ind: London, 23: 958-959. |
| [13] | Buttery R G, Ling L C, Juliano B O, Turnbaugh J G. 1983. Cooked rice aroma and 2-acetyl-1-pyrroline. J Agric Food Chem, 31(4): 823-826. |
| [14] | Champagne E T, Bett-Garber K L, Thompson J, Mutters R, Grimm C C, McClung A M. 2005. Effects of drain and harvest dates on rice sensory and physicochemical properties. Cereal Chem, 82(4): 369-374. |
| [15] | Champagne E T, Bett-Garber K L, Grimm C C, McClung A M. 2007. Effects of organic fertility management on physicochemical properties and sensory quality of diverse rice cultivars. Cereal Chem, 84(4): 320-327. |
| [16] | Champagne E T. 2008. Rice aroma and flavor: A literature review. Cereal Chem, 85(4): 445-454. |
| [17] | Cha-um S, Vejchasarn P, Kirdmanee C. 2007. An effective defensive response in Thai aromatic rice varieties (Oryza sativa L. spp. indica) to salinity. J Crop Sci Biotechnol, 10(4): 257-264. |
| [18] | Chen M L, Wei X J, Shao G N, Tang S Q, Luo J, Hu P S. 2012. Fragrance of the rice grain achieved via artificial microRNA- induced down-regulation of OsBADH2. Plant Breeding, 131(5): 584-590. |
| [19] | Chen M S, Presting G, Barbazuk W B, Goicoechea J L, Blackmon B, Fang G C, Kim H, Frisch D, Yu Y, Sun S, Higingbottom S, Phimphilai J, Phimphilai D, Thurmond S, Barnett L, Costa R, Williams B, Walser S, Atkins M, Hall C, Budiman M A, Tomkins J P, Luo M Z, Bancroft I, Salse J, Regad F, Mohapatra T, Singh N K, Tyagi A K, Soderlund C, Dean R A, Wing R A. 2002. An integrated physical and genetic map of the rice genome. Plant Cell, 14(3): 537-545. |
| [20] | Chen S H, Yang Y, Shi W W, Ji Q, He F, Zhang Z D, Cheng Z K, Liu X N, Xu M L. 2008. Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell, 20(7): 1850-1861. |
| [21] | Efferson J N. 1985. Rice quality in world markets. In: IRRI. Rice Grain Quality and Marketing. Manila, the Philippine: International Rice Research Institute: 1-13. |
| [22] | Feng H Y, Jiang H L, Wang M, Tang X R, Duan M Y, Pan S G, Tian H, Wang S L, Mo Z W. 2019. Morphophysiological responses of different scented rice varieties to high temperature at seedling stage. Chin J Rice Sci, 33(1): 68-74. (in Chinese with English abstract) |
| [23] | Fiaz S, Ahmad S, Noor M A, Wang X K, Younas A, Riaz A, Riaz A, Ali F. 2019. Applications of the CRISPR/Cas9 system for rice grain quality improvement: Perspectives and opportunities. Int J Mol Sci, 20(4): 888. |
| [24] | Fitzgerald T L, Waters D L E, Henry R J. 2008. The effect of salt on betaine aldehyde dehydrogenase transcript levels and 2-acetyl-1-pyrroline concentration in fragrant and non-fragrant rice (Oryza sativa, L.). Plant Sci, 175(4): 539-546. |
| [25] | Fitzgerald T L, Waters D L E, Brooks L O, Henry R J. 2010. Fragrance in rice (Oryza sativa) is associated with reduced yield under salt treatment. Environ Exp Bot, 68(3): 292-300. |
| [26] | Gaur A, Wani S, Pandita D, Bharti N, Malav A, Shikari A, Bhat A. 2016. Understanding the fragrance in rice. J Rice Res, 4(1): e125. |
| [27] | Gay F, Maraval I, Roques S, Gunata Z, Boulanger R, Audebert A, Mestres C. 2010. Effect of salinity on yield and 2-acetyl-1- pyrroline content in the grains of three fragrant rice cultivars (Oryza sativa L.) in Camargue (France). Field Crops Res, 117(1): 154-160. |
| [28] | Goufo P, Duan M, Wongpornchai S, Tang X. 2010. Some factors affecting the concentration of the aroma compound 2-acetyl-1- pyrroline in two fragrant rice cultivars grown in South China. Front Agric China, 4(1): 1-9. |
| [29] | Hashemi F S G, Rafii M Y, Ismail M R, Mahmud T M M, Rahim H A, Asfaliza R, Malek M A, Latif M A. 2013. Biochemical, genetic and molecular advances of fragrance characteristics in rice. Crit Rev Plant Sci, 32(6): 445-457. |
| [30] | Hashemi F S G, Ismail M R, Rafii M Y, Aslani F, Miah G, Muharam F M. 2018. Critical multifunctional role of the betaine aldehyde dehydrogenase gene in plants. Biotechnol Biotechnol Equip, 32(4): 815-829. |
| [31] | Hasthanasombut S, Ntui V, Supaibulwatana K, Mii M, Nakamura I. 2010. Expression of indica rice OsBADH1 gene under salinity stress in transgenic tobacco. Plant Biotechnol Rep, 4(1): 75-83. |
| [32] | Hasthanasombut S, Supaibulwatana K, Mii M, Nakamura I. 2011. Genetic manipulation of japonica rice using the OsBADH1 gene from indica rice to improve salinity tolerance. Plant Cell Tissue Organ Cult, 104(1): 79-89. |
| [33] | He Q, Park Y J. 2015. Discovery of a novel fragrant allele and development of functional markers for fragrance in rice. Mol Breeding, 35(11): 217. |
| [34] | Hinge V R, Patil H B, Nadaf A B. 2016. Aroma volatile analyses and 2AP characterization at various developmental stages in Basmati and non-Basmati scented rice (Oryza sativa L.) cultivars. Rice, 9(1): 38. |
| [35] | Hong Z L, Lakkineni K, Zhang Z M, Verma D P S. 2000. Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol, 122(4): 1129-1136. |
| [36] | Hoque T S, Hossain M A, Mostofa M G, Burritt D J, Fujita M, Tran L S P. 2016. Methylglyoxal: An emerging signaling molecule in plant abiotic stress responses and tolerance. Front Plant Sci, 7: 1341. |
| [37] | Huang T C, Teng C S, Chang J L, Chuang H S, Ho C T, Wu M L. 2008. Biosynthetic mechanism of 2-acetyl-1-pyrroline and its relationship with Δ1-pyrroline-5-carboxylic acid and methylglyoxal in aromatic rice (Oryza sativa L.) callus. J Agric Food Chem, 56(16): 7399-7404. |
| [38] | IRGSP (International Rice Genome Sequencing Project), Sasaki T. 2005. The map-based sequence of the rice genome. Nature, 436: 793-800. |
| [39] | Itani T, Tamaki M, Hayata Y, Fushimi T, Hashizume K. 2004. Variation of 2-acetyl-1-pyrroline concentration in aromatic rice grains collected in the same region in Japan and factors affecting its concentration. Plant Prod Sci, 7(2): 178-183. |
| [40] | Jin Q S, Waters D, Cordeiro G M, Henry R J, Reinke R F. 2003. A single nucleotide polymorphism (SNP) marker linked to the fragrance gene in rice (Oryza sativa L.). Plant Sci, 165(2): 359-364. |
| [41] | Kaikavoosi K, Kad T D, Zanan R L, Nadaf A B. 2015. 2-acetyl-1- pyrroline augmentation in scented indica rice (Oryza sativa L.) varieties through Δ1-pyrroline-5-carboxylate synthetase (P5CS) gene transformation. Appl Biochem Biotechnol, 177(7): 1466-1479. |
| [42] | Khandagale K S, Chavan R, Nadaf A B. 2017. RNAi mediated down regulation of BADH2 gene for expression of 2-acetyl-1- pyrroline in non-scented indica rice IR-64 (Oryza sativa L.). Can J Biotechnol, 1: 169. |
| [43] | Kinnersley A M, Turano F J. 2000. Gamma aminobutyric acid (GABA) and plant responses to stress. Crit Rev Plant Sci, 19(6): 479-509. |
| [44] | Kovach M J, Calingacion M N, Fitzgerald M A, McCouch S R. 2009. The origin and evolution of fragrance in rice (Oryza sativa L.). Proc Natl Acad Sci USA, 106(34): 14444-14449. |
| [45] | Lorieux M, Petrov M, Huang N, Guiderdoni E, Ghesquière A. 1996. Aroma in rice: Genetic analysis of a quantitative trait. Theor Appl Genet, 93(7): 1145-1151. |
| [46] | Mahajan G, Matloob A, Singh R, Singh V P, Chauhan B S. 2018. Basmati rice in the Indian subcontinent: Strategies to boost production and quality traits. Adv Agron, 151: 159-213. |
| [47] | Maraval I, Sen K, Agrebi A, Menut C, Morere A, Boulanger R, Gay F, Mestres C, Gunata Z. 2010. Quantification of 2-acetyl-1- pyrroline in rice by stable isotope dilution assay through headspace solid-phase microextraction coupled to gas chromatography- tandem mass spectrometry. Anal Chim Acta, 675(2): 148-155. |
| [48] | Mitsuya S, Yokota Y, Fujiwara T, Mori N, Takabe T. 2009. OsBADH1 is possibly involved in acetaldehyde oxidation in rice plant peroxisomes. FEBS Lett, 583(22): 3625-3629. |
| [49] | Mo Z W, Li W, Pan S G, Fitzgerald T L, Xiao F, Tang Y J, Wang Y L, Duan M Y, Tian H, Tang X R. 2015. Shading during the grain filling period increases 2-acetyl-1-pyrroline content in fragrant rice. Rice, 8(1): 9. |
| [50] | Mo Z W, Huang J X, Xiao D, Ashraf U, Duan M Y, Pan S G, Tian H, Xiao L Z, Zhong K Y, Tang X R. 2016. Supplementation of 2-Ap, Zn and La improves 2-acetyl-1-pyrroline concentrations in detached aromatic rice panicles in vitro. PLoS One, 11(2): e0149523. |
| [51] | Monggoot S, Sookwong P, Mahatheeranont S, Meechoui S. 2014. Influence of single nutrient element on 2-acetyl-1-pyrroline contents in Thai fragrant rice (Oryza sativa L.) cv. Khao DawkMali 105 grown under soilless conditions. In: Proceedings of the 26th Annual Meeting of the Thai Society for Biotechnology and International Conference. Mae Fah Luang University Chiang Rai: 642-647. |
| [52] | Nadaf A B, Wakte K V, Zanan R L. 2014. 2-acetyl-1-pyrroline biosynthesis: From fragrance to a rare metabolic disease. J Plant Sci Res, 1: 102. |
| [53] | Niu X L, Zheng W J, Lu B R, Ren G J, Huang W Z, Wang S H, Liu J L, Tang Z Z, Luo D, Wang Y G, Liu Y S. 2007. An unusual posttranscriptional processing in two betaine aldehyde dehydrogenase loci of cereal crops directed by short, direct repeats in response to stress conditions. Plant Physiol, 143(4): 1929-1942. |
| [54] | Niu X L, Tang W, Huang W Z, Ren G J, Wang Q L, Luo D, Xiao Y Y, Yang S M, Wang F, Lu B R, Gao F Y, Lu T G, Liu Y S. 2008. RNAi-directed down regulation of OsBADH2 results in aroma (2-acetyl-1-pyrroline) production in rice (Oryza sativa L.). BMC Plant Biol, 8(1): 100. |
| [55] | Okpala N E, Mo Z W, Duan M Y, Tang X R. 2018. The genetics and biosynthesis of 2-acetyl-1-pyrroline in fragrant rice. Plant Physiol Biochem, 135: 272-276. |
| [56] | Pachauri V, Singh M K, Singh A K, Singh S, Shakeel N A, Singh V P, Singh N K. 2010. Origin and genetic diversity of aromatic rice varieties, molecular breeding and chemical and genetic basis of rice aroma. J Plant Biochem Biotechnol, 19(2): 127-143. |
| [57] | Pachauri V, Mishra V, Mishra P, Singh A K, Singh S, Singh R, Singh N K. 2014. Identification of candidate genes for rice grain aroma by combining QTL mapping and transcriptome profiling approaches. Cereal Res Commun, 42(3): 376-388. |
| [58] | Peng B, Zuo Y H, Hao Y L, Peng J, Kong D Y, Peng Y, Nassiron T Y, He L L, Sun Y F, Liu L, Pang R H, Chen Y X, Li J T, Zhou Q Y, Duan B, Song X H, Song S Z, Yuan H Y. 2018. Studies on aroma gene and its application in rice genetics and breeding. J Plant Stud, 7(2): 29-41. |
| [59] | Phillips S A, Thornalley P J. 1993. The formation of methylglyoxal from triose phosphates: Investigation using a specific assay for methylglyoxal. Eur J Biochem, 212(1): 101-105. |
| [60] | Poonlaphdecha J, Maraval I, Roques S, Audebert A, Boulanger R, Bry X, Gunata Z. 2012. Effect of timing and duration of salt treatment during growth of a fragrant rice variety on yield and 2-acetyl-1-pyrroline, proline, and GABA levels. J Agric Food Chem, 60(15): 3824-3830. |
| [61] | Prathepha P. 2009. The fragrance (fgr) gene in natural populations of wild rice (Oryza rufipogon Griff.). Genet Resour Crop Evol, 56(1): 13-18. |
| [62] | Prodhan Z H, Faruq G, Rashid K A, Taha R M. 2017. Effects of temperature on volatile profile and aroma quality in rice. Int J Agric Biol, 19(5): 1065-1072. |
| [63] | Romanczyk Jr L J, McClelland C A, Post L S, Aitken W M. 1995. Formation of 2-acetyl-1-pyrroline by several Bacillus cereus strains isolated from cocoa fermentation boxes. J Agric Food Chem, 43(2): 469-475. |
| [64] | Sakthivel K, Sundaram R M, Shobha Rani N, Balachandran S M, Neeraja C N. 2009. Genetic and molecular basis of fragrance in rice. Biotechnol Adv, 27(4): 468-473. |
| [65] | Shan Q W, Wang Y P, Chen K L, Liang Z, Li J, Zhang Y, Zhang K, Liu J X, Voytas D F, Zheng X L, Zhang Y, Gao C X. 2013. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol Plant, 6(4): 1365-1368. |
| [66] | Shan Q W, Zhang Y, Chen K L, Zhang K, Gao C X. 2015. Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol J, 13(6): 791-800. |
| [67] | Shao G N, Xie L H, Jiao G A, Wei X J, Sheng Z H, Tang S Q, Hu P S. 2017. CRISPR/Cas9-mediated editing of the fragrant gene Badh2 in rice. Chin J Rice Sci, 31(2): 216-222. (in Chinese with English abstract) |
| [68] | Shelp B J, Bozzo G G, Trobacher C P, Zarei A, Deyman K L, Brikis C J. 2012. Hypothesis/review: Contribution of putrescine to 4-aminobutyrate (GABA) production in response to abiotic stress. Plant Sci, 193: 130-135. |
| [69] | Shi W W, Yang Y, Chen S H, Xu M L. 2008. Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol Breeding, 22(2): 185-192. |
| [70] | Shirasawa K, Takabe T, Takabe T, Kishitani S. 2006. Accumulation of glycinebetaine in rice plants that overexpress choline monooxygenase from spinach and evaluation of their tolerance to abiotic stress. Ann Bot, 98(3): 565-571. |
| [71] | Siddiq E A, Vemireddy L R, Nagaraju J. 2012. Basmati rices: Genetics, breeding and trade. Agric Res, 1(1): 25-36. |
| [72] | Singh A, Singh P K, Singh R, Pandit A, Mahato A K, Gupta D K, Tyagi K, Singh A K, Singh N K, Sharma T R. 2010. SNP haplotypes of the BADH1 gene and their association with aroma in rice (Oryza sativa L.). Mol Breeding, 26(2): 325-338. |
| [73] | Singh R, Singh A K, Sharma T R, Singh A, Singh N K. 2007. Fine mapping of aroma QTLs in Basmati rice (Oryza sativa L.) on chromosomes 3, 4 and 8. J Plant Biochem Biotechnol, 16(2): 75-82. |
| [74] | Singh R K, Singh U S, Khush G S, Rohilla R. 2000. Genetics and biotechnology of quality traits in aromatic rices. In: Singh R K, Singh U S, Khush G S. Aromatic Rices. Manila, the Philippines: International Rice Research Institute: 47-70. |
| [75] | Somta P, Kuswanto K, Srinives P. 2019. The genetics of pandan- like fragrance, 2-acetyl-1-pyrroline, in crops. AGRIVITA, J Agric Sci, 41(1): 10-22. |
| [76] | Srinivas P, Sulochanamma G, Raghavan B, Gurudutt K. 2006. Process for the stabilization of 2-acetyl-1pyrroline, the basmati rice flavourant: US, PAT-US2006147596 2006-07-06.. |
| [77] | Talukdar P R, Rathi S, Pathak K, Chetia S K, Sarma R N. 2017. Population structure and marker-trait association in indigenous aromatic rice. Rice Sci, 24(3): 145-154. |
| [78] | Tester M, Davenport R. 2003. Na+ tolerance and Na+ transport in higher plants. Ann Bot, 91(5): 503-527. |
| [79] | Vanavichit A, Yoshihashi T. 2010. Molecular aspects of fragrance and aroma in rice. Adv Bot Res, 56: 49-73. |
| [80] | Wakte K, Zanan R, Hinge V, Khandagale K, Nadaf A, Henry R. 2017. Thirty-three years of 2-acetyl-1-pyrroline, a principal basmati aroma compound in scented rice (Oryza sativa L.): A status review. J Sci Food Agric, 97(2): 384-395. |
| [81] | Wang P, Tang X R, Tian H, Pan S G, Duan M Y, Nie J, Luo Y M, Xiao L Z. 2013. Effects of different irrigation modes on aroma content of aromatic rice at booting stage. Guangdong Agric Sci, 8: 1-3. (in Chinese with English abstract) |
| [82] | Widjaja R, Craske J D, Wootton M. 1996. Comparative studies on volatile components of non-fragrant and fragrant rices. J Sci Food Agric, 70(2): 151-161. |
| [83] | Wijerathna Y M A M, Kottearachchi N S, Gimhani D R, Sirisena D N. 2014. Exploration of relationship between fragrant gene and growth performances of fragrant rice (Oryza sativa L.) seedlings under salinity stress. J Exp Biol Agric Sci, 2(1): 7-12. |
| [84] | Wu M L, Chou K L, Wu C R, Chen J K, Huang T C. 2009. Characterization and the possible formation mechanism of 2-acetyl-1-pyrroline in aromatic vegetable soybean (Glycine max L.). J Food Sci, 74(5): S192-S197. |
| [85] | Yang D S, Shewfelt R L, Lee K S, Kays S J. 2008. Comparison of odor-active compounds from six distinctly different rice flavor types. J Agric Food Chem, 56(8): 2780-2787. |
| [86] | Yi M, Nwe K T, Vanavichit A, Chai-arree W, Toojinda T. 2009. Marker assisted backcross breeding to improve cooking quality traits in Myanmar rice cultivar Manawthukha. Field Crops Res, 113(2): 178-186. |
| [87] | Yoshihashi T, Huong N T T, Inatomi H. 2002. Precursors of 2-acetyl-1-pyrroline, a potent flavor compound of an aromatic rice variety. J Agric Food Chem, 50(7): 2001-2004. |
| [88] | Yoshihashi T, Nguyen T T H, Kabaki N. 2004. Area dependency of 2-acetyl-1-pyrroline content in an aromatic rice variety, Khao Dawk Mali 105. Jpn Agric Res Q, 38(2): 105-109. |
| [89] | Yoshihashi T, Huong N T T, Surojanametakul V, Tungtrakul P, Varanyanond W. 2005. Effect of storage conditions on 2-acetyl-1-pyrroline content in aromatic rice variety, Khao Dawk Mali 105. J Food Sci, 70(1): S34-S37. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||