
Rice Science ›› 2025, Vol. 32 ›› Issue (4): 575-584.DOI: 10.1016/j.rsci.2025.01.005
• • 上一篇
收稿日期:2024-10-14
接受日期:2025-01-16
出版日期:2025-07-28
发布日期:2025-08-06
. [J]. Rice Science, 2025, 32(4): 575-584.
| Primer set | Primer name | Primer and probe sequence (5ʹ‒3ʹ) | Amplicon length (bp) |
|---|---|---|---|
| P1 | RAAF1 | GCCGCTAGGAATGAGCAATGCACACGTGGA | 162 |
| RAAR1 | GCGTCCTCGTCTAAGCGATAGACGGCGCAC | ||
| P2 | RAAF2 | CACACGTGGAAAGGGACCTCAAAAAAATCC | 158 |
| RAAR2 | CTTGCAAGGGATAGAAGCGTCCTCGTCTAA | ||
| P3 | RAAF3 | CCGTGCTGTGTGGAGTACAAAGCCACCAAC | 131 |
| RAAR3 | ATAACTTGTGACGTAGCGAGCGTTTGAGAT | ||
| P4 | RAAF4 | GGTGCAACAAACTCCCATGGTGTGACGGG | 134 |
| RAAR4 | CAGTCCGGATTGGAGTCTGCAACTCGACTC | ||
| P5 | RAAF5 | ACCCAACATCCAGTTCGCATCGTTTAGGGC | 221 |
| RAAR5 | CTCAACCTGGGAATTGCAGTGGATACTGGG | ||
| / | RAAR2-biotin | Biotin-CTTGCAAGGGATAGAAGCGTCCTCGTCTAA | / |
| / | RAA2-nfo-probe | FAM-TAGTGCGCCAGGAAGGCCAGCGCACCGTAGTTGC-THF-GACGGCTACCACCGT (C3 space) | / |
Table 1. Primers and probes for recombinase-aided amplification-lateral flow dipstick (RAA-LFD) detection of Xanthomonas oryzae pv. oryzae.
| Primer set | Primer name | Primer and probe sequence (5ʹ‒3ʹ) | Amplicon length (bp) |
|---|---|---|---|
| P1 | RAAF1 | GCCGCTAGGAATGAGCAATGCACACGTGGA | 162 |
| RAAR1 | GCGTCCTCGTCTAAGCGATAGACGGCGCAC | ||
| P2 | RAAF2 | CACACGTGGAAAGGGACCTCAAAAAAATCC | 158 |
| RAAR2 | CTTGCAAGGGATAGAAGCGTCCTCGTCTAA | ||
| P3 | RAAF3 | CCGTGCTGTGTGGAGTACAAAGCCACCAAC | 131 |
| RAAR3 | ATAACTTGTGACGTAGCGAGCGTTTGAGAT | ||
| P4 | RAAF4 | GGTGCAACAAACTCCCATGGTGTGACGGG | 134 |
| RAAR4 | CAGTCCGGATTGGAGTCTGCAACTCGACTC | ||
| P5 | RAAF5 | ACCCAACATCCAGTTCGCATCGTTTAGGGC | 221 |
| RAAR5 | CTCAACCTGGGAATTGCAGTGGATACTGGG | ||
| / | RAAR2-biotin | Biotin-CTTGCAAGGGATAGAAGCGTCCTCGTCTAA | / |
| / | RAA2-nfo-probe | FAM-TAGTGCGCCAGGAAGGCCAGCGCACCGTAGTTGC-THF-GACGGCTACCACCGT (C3 space) | / |
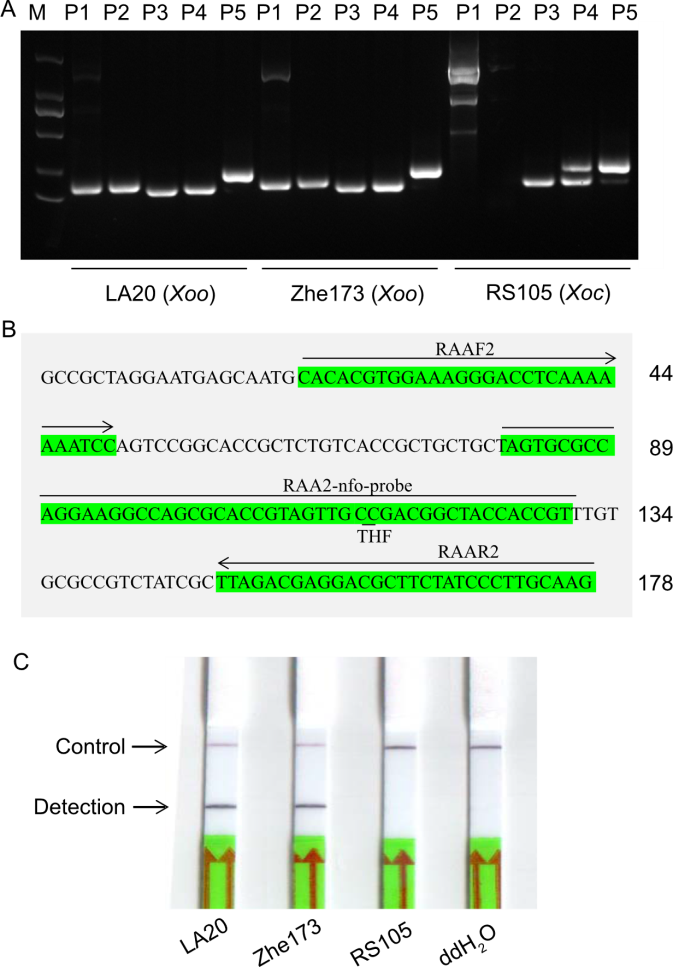
Fig. 1. Primers and probe screening for recombinase-aided amplification-lateral flow dipstick (RAA-LFD) detection of Xoo. A, Primer screening. B, Position of selected primer (RAAF2/R2) and probe (RAA2-nfo-probe) based on the Xoo locus XooORF0080. C, RAA-LFD results using primers (RAAF2/R2) and probe (RAA2-nfo-probe). M, 2000 bp DNA marker (TaKaRa, Japan); P1‒P5, The designed primer pairs RAAF1/R1, RAAF2/R2, RAAF3/R3, RAAF4/R4, and RAAF5/R5, respectively; Xoo, Xanthomonas oryzae pv. oryzae; Xoc, Xanthomonas oryzae pv. oryzicola; LA20 and Zhe173 are Xoo strains, and RS105 is a Xoc strain. ddH2O was used as a negative control.
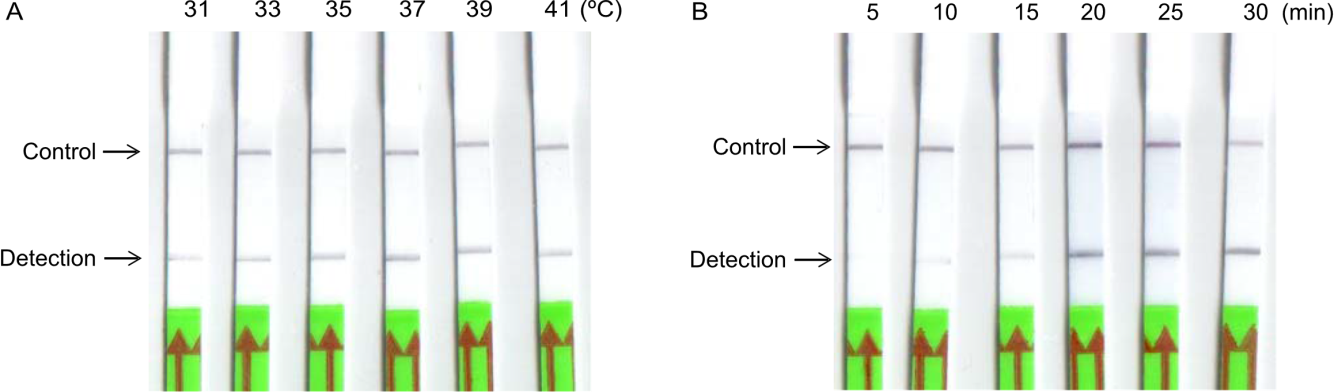
Fig. 2. Optimization of reaction temperature and time for recombinase-aided amplification-lateral flow dipstick (RAA-LFD) detection of Xanthomonas oryzae pv. oryzae. A and B, Reaction temperature (from 31 ºC to 41 ºC for 20 min, A) and time (from 5 to 30 min at 37 ºC, B) optimization.
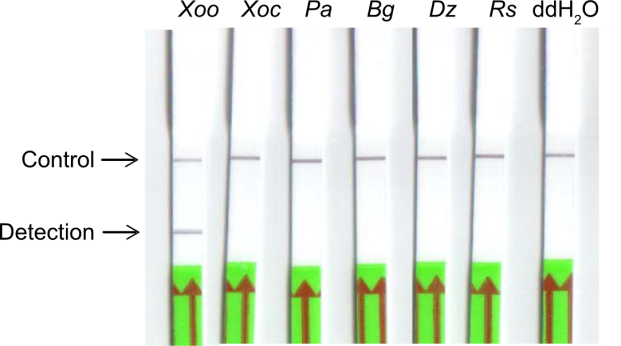
Fig. 3. Specificity assessment for recombinase-aided amplification lateral flow dipstick (RAA-LFD) detection of Xoo. Xoo, Xanthomonas oryzae pv. oryzae; Xoc, Xanthomonas oryzae pv. oryzicola; Pa, Pantoea ananatis; Bg, Burkholderia glumae; Dz, Dickeya zeae; Rs, Ralstonia solanacearum. ddH2O was used as a negative control.
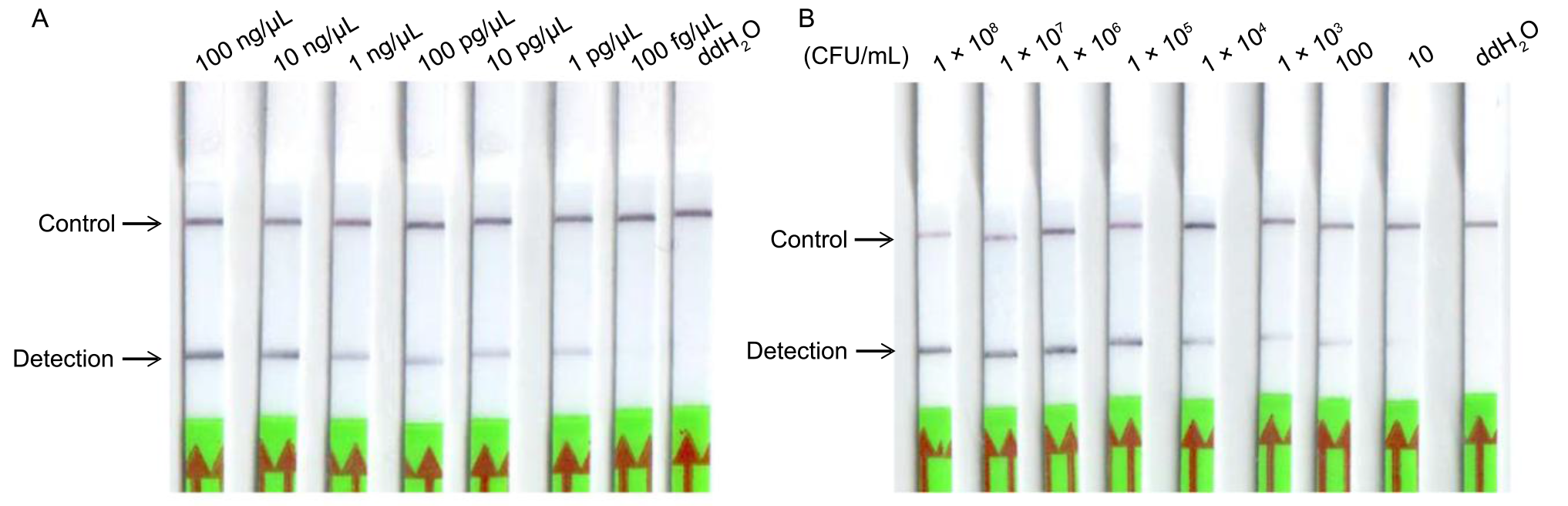
Fig. 4. Sensitivity assessment for recombinase-aided amplification-lateral flow dipstick (RAA-LFD) detection of Xanthomonas oryzae pv. oryzae (Xoo). A, Sensitivity assessment was determined using Xoo genomic DNA as the template, with concentrations ranging from 100 ng/μL to 100 fg/μL. B, Sensitivity assessment was performed using Xoo culture solution as the template, with concentrations ranging from 1 × 108 CFU/mL to 10 CFU/mL. ddH2O was used as a negative control.
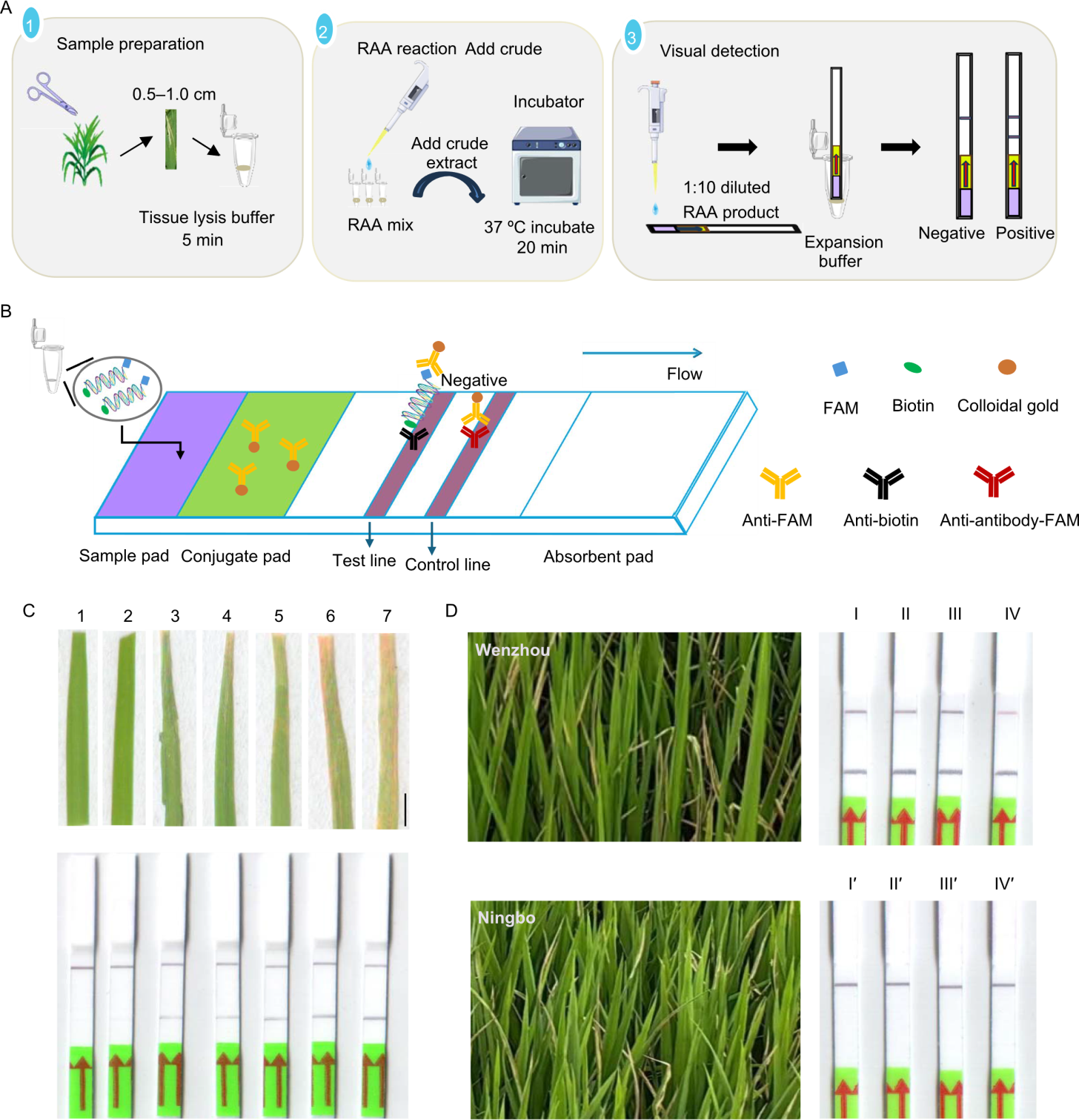
Fig. 5. On-site detection of Xanthomonas oryzae pv. oryzae (Xoo) by developed recombinase-aided amplification-lateral flow dipstick (RAA-LFD) system. A, Workflow of RAA-LFD detection: sample preparation with simplified DNA extraction, RAA reaction at 37 ºC for 20 min, and visual detection by LFD. B, Schematic diagram of LFD detection. FAM (carboxyfluorescein amidite) of special amplicons is first caputured by anti-FAM antibody conjuated colloidal gold in conjuated pad. Then, biotin of the complex binds to anti-biotin antibody in the test line, leading to appearing test line, indicating positive, otherwise indicating negative. The complex of anti-FAM antibody conjuated colloidal gold binds to anti-antibody-FAM antibody in the control line, leading to appearing the control line. C, RAA-LFD detection result for the inoculated rice leaves. Lanes 1‒7, 1‒7 d after inoculation. Rice variety Nipponbare was used. Scale bar, 5 mm. D, RAA-LFD detection result for the field samples suspected to be infected by Xoo from Wenzhou and Ningbo cities, Zhejiang Province, China. Lanes I‒IV, Leaf samples from Wenzhou with lesion lengths categorized as less than 1 cm, 1‒3 cm, 3‒6 cm, and greater than 6 cm, respectively; Lanes Iʹ‒IVʹ, Leaf samples from Ningbo with corresponding lesion lengths of less than 1 cm, 1‒3 cm, 3‒6 cm, and greater than 6 cm, respectively.
| Geographic location | Number of samples based on lession length | RAA-LFD | |||||
|---|---|---|---|---|---|---|---|
| < 1 cm | 1‒3 cm | 3‒6 cm | > 6 cm | Positive | Negative | ||
| Wenzhou | 6 | 6 | 4 | 2 | 18 | 0 | |
| Ningbo | 6 | 5 | 3 | 1 | 0 | 15 | |
| Total | 12 | 11 | 7 | 3 | 18 | 15 | |
Table 2. Recombinase-aided amplification-lateral flow dipstick (RAA-LFD) detection results of field samples.
| Geographic location | Number of samples based on lession length | RAA-LFD | |||||
|---|---|---|---|---|---|---|---|
| < 1 cm | 1‒3 cm | 3‒6 cm | > 6 cm | Positive | Negative | ||
| Wenzhou | 6 | 6 | 4 | 2 | 18 | 0 | |
| Ningbo | 6 | 5 | 3 | 1 | 0 | 15 | |
| Total | 12 | 11 | 7 | 3 | 18 | 15 | |
| [1] | Amaral C, Antunes W, Moe E L, et al. 2021. A molecular test based on RT-LAMP for rapid, sensitive and inexpensive colorimetric detection of SARS-CoV-2 in clinical samples. Sci Rep, 11(1): 16430. |
| [2] | Awaludin N, Abdullah J, Salam F, et al. 2020. Fluorescence-based immunoassay for the detection of Xanthomonas oryzae pv. oryzae in rice leaf. Anal Biochem, 610: 113876. |
| [3] | Bai Y H, Wu X H, Liu J J, et al. 2024. Establishment and application prospect of reverse transcriptase recombinase-aided amplification assay for subgroup C avian metapneumovirus. Vet Sci, 11(3): 122. |
| [4] | Buddhachat K, Ritbamrung O, Sripairoj N, et al. 2021. One-step colorimetric LAMP (cLAMP) assay for visual detection of Xanthomonas oryzae pv. oryzae in rice. Crop Prot, 150: 105809. |
| [5] | Buddhachat K, Sripairoj N, Ritbamrung O, et al. 2022. RPA-assisted Cas12a system for detecting pathogenic Xanthomonas oryzae, a causative agent for bacterial leaf blight disease in rice. Rice Sci, 29(4): 340-352. |
| [6] | Buddhachat K, Ritbamrung O, Inthima P, et al. 2024. Rapid detection of two pathogenically important Xanthomonas in rice using a loop-mediated isothermal amplification with lateral flow dipstick (LAMP-LFD). Crop Prot, 175: 106466. |
| [7] | Cao Y H, Song X M. 2023. Meat authenticity made easy: DNA extraction-free rapid onsite detection of duck and pork ingredients in beef and lamb using dual-recombinase-aided amplification and multiplex lateral flow strips. J Agric Food Chem, 71: 14782-14794. |
| [8] | Cao Y H, Weng H T, Rao S F, et al. 2023. Rapid and visual field diagnosis of tomato brown rugose fruit virus using reverse transcription recombinase aided amplification (RT RAA) combined with lateral flow strips (LFS). Crop Prot, 173: 106355. |
| [9] | Cui Z, Ojaghian M R, Tao Z, et al. 2016. Multiplex PCR assay for simultaneous detection of six major bacterial pathogens of rice. J Appl Microbiol, 120(5): 1357-1367. |
| [10] | Ding X Y, Wang H J, Cui M Q, et al. 2022. Development of a real-time recombinase-aided amplification method to rapidly detect methicillin-resistant Staphylococcus aureus. Microorganisms, 10(12): 2351. |
| [11] | Fang W W, Cai Y Y, Zhu L, et al. 2021. Rapid and highly sensitive detection of toxigenic Vibrio cholerae based on recombinase-aided amplification combining with lateral flow assay. Food Anal Meth, 14(4): 687-696. |
| [12] | Guo Z H, Xing G X, Li P, et al. 2023. Development and application of a recombinase-aided amplification and lateral flow assay for rapid detection of pseudorabies virus from clinical crude samples. Int J Biol Macromol, 224: 646-652. |
| [13] | Hou Y X, Wang L Y, Wang L, et al. 2015. JMJ704 positively regulates rice defense response against Xanthomonas oryzae pv. oryzae infection via reducing H3K4me2/3 associated with negative disease resistance regulators. BMC Plant Biol, 15: 286. |
| [14] | Hou Y X, Xu Y, Zhang Y, et al. 2020. First report of bacterial panicle blight of rice caused by Burkholderia glumae in Southern China. Plant Dis, 104(4): 1252. |
| [15] | Hou Y X, Liang Y, Yang C D, et al. 2023. Complete genomic sequence of Xanthomonas oryzae pv. oryzae strain, LA20, for studying resurgence of rice bacterial blight in the Yangtze River region, China. Int J Mol Sci, 24(9): 8132. |
| [16] | Hu Y F, Pan F J, You J, et al. 2023. First report of Aphelenchoides besseyi causing white tip disease of rice in Heilongjiang Province of China. Plant Dis, 107(7): 2264. |
| [17] | Ju Y H, Tian H J, Zhang R H, et al. 2017. Overexpression of OsHSP18.0-CI enhances resistance to bacterial leaf streak in rice. Rice, 10(1): 12. |
| [18] | Kim S H, Lee S Y, Kim U, et al. 2023. Diverse methods of reducing and confirming false-positive results of loop-mediated isothermal amplification assays: A review. Anal Chim Acta, 1280: 341693. |
| [19] | Lang J M, Hamilton J P, Diaz M G Q, et al. 2010. Genomics-based diagnostic marker development for Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola. Plant Dis, 94(3): 311-319. |
| [20] | Lang J M, Langlois P, Nguyen M H R, et al. 2014. Sensitive detection of Xanthomonas oryzae pathovars oryzae and oryzicola by loop-mediated isothermal amplification. Appl Environ Microbiol, 80(15): 4519-4530. |
| [21] | Li Q, Duan L J, Jin D S, et al. 2024. A real-time fluorogenic recombinase polymerase amplification microfluidic chip (on-chip RPA) for multiple detection of pathogenic microorganisms of penaeid shrimp. Aquaculture, 578: 740017. |
| [22] | Li X P, Zhu S Y, Zhang X L, et al. 2023. Advances in the application of recombinase-aided amplification combined with CRISPR-Cas technology in quick detection of pathogenic microbes. Front Bioeng Biotechnol, 11: 1215466. |
| [23] | Li Y D, Yu Z R, Jiao S L, et al. 2020. Development of a recombinase-aided amplification assay for rapid and sensitive detection of porcine circovirus 3. J Virol Methods, 282: 113904. |
| [24] | Lin H, Zhao S, Liu Y H, et al. 2022. Rapid visual detection of Plasmodium using recombinase-aided amplification with lateral flow dipstick assay. Front Cell Infect Microbiol, 12: 922146. |
| [25] | Liu D, Shen H C, Zhang Y Q, et al. 2021. A microfluidic-integrated lateral flow recombinase polymerase amplification (MI-IF-RPA) assay for rapid COVID-19 detection. Lab Chip, 21(10): 2019-2026. |
| [26] | Liu J, Qian W D, Wang J, et al. 2023. A recombinase-aided amplification assay for the detection of Chlamydia felis. Pol J Microbiol, 72(3): 339-343. |
| [27] | Lu C C, Hao M X, Kong L G, et al. 2022. First report of new bacterial leaf streak of rice caused by Pantoea ananatis in China. Plant Dis, 106(2): 1747. |
| [28] | Lu W, Pan L Q, Zhao H J, et al. 2014. Molecular detection of Xanthomonas oryzae pv. oryzae, Xanthomonas oryzae pv. oryzicola, and Burkholderia glumae in infected rice seeds and leaves. Crop J, 2( 6): 398-406. |
| [29] | Luna E, Lang J, McClung A, et al. 2023. First report of rice bacterial leaf blight disease caused by Pantoea ananatis in the United States. Plant Dis, 107(7): 2214. |
| [30] | Munawar M A. 2022. Critical insight into recombinase polymerase amplification technology. Expert Rev Mol Diagn, 22(7): 725-737. |
| [31] | Niño-Liu D O, Ronald P C, Bogdanove A J. 2006. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol Plant Pathol, 7(5): 303-324. |
| [32] | Ou S Q, Gao J, Peng D L, et al. 2014. First report of Aphelenchoides besseyi causing white tip disease of rice in Jilin Province, China. Plant Dis, 98(8): 1165. |
| [33] | Pang F, Long Q Q. 2023. Recent advances in diagnostic approaches for orf virus. Appl Microbiol Biotechnol, 107(5/6): 1515-1523. |
| [34] | Park J W. 2022. Principles and applications of loop-mediated isothermal amplification to point-of-care tests. Biosensors, 12(10): 857. |
| [35] | Piepenburg O, Williams C H, Stemple D L, et al. 2006. DNA detection using recombination proteins. PLoS Biol, 4(7): e204. |
| [36] | Qi Z Q, Ju F Y, Guo Y X, et al. 2024. A rapid, equipment-free method for detecting avirulence genes of Pyricularia oryzae using a lateral flow strip-based RPA assay. Plant Dis, 108(8): 2283-2290. |
| [37] | Sakthivel N, Mortensen C N, Mathur S B. 2001. Detection of Xanthomonas oryzae pv. oryzae in artificially inoculated and naturally infected rice seeds and plants by molecular techniques. Appl Microbiol Biotechnol, 56(3/4): 435-441. |
| [38] | Shen X X, Qiu F Z, Shen L P, et al. 2019. A rapid and sensitive recombinase aided amplification assay to detect hepatitis B virus without DNA extraction. BMC Infect Dis, 19(1): 229. |
| [39] | Song E S, Kim S Y, Noh T H, et al. 2014. PCR-based assay for rapid and specific detection of the new Xanthomonas oryzae pv. oryzae K3a race using an AFLP-derived marker. J Microbiol Biotechnol, 24(6): 732-739. |
| [40] | Soroka M, Wasowicz B, Rymaszewska A. 2021. Loop-mediated isothermal amplification (LAMP): The better sibling of PCR? Cells, 10(8): 1931. |
| [41] | Tan M Y, Liao C, Liang L N, et al. 2022. Recent advances in recombinase polymerase amplification: Principle, advantages, disadvantages and applications. Front Cell Infect Microbiol, 12: 1019071. |
| [42] | Wang A Y, Luo J, Wang C L, et al. 2023. Establishment and application of the Recombinase-Aided Amplification-Lateral Flow Dipstick detection method for Pantoea ananatis on rice. Austral Plant Pathol, 52: 283-291. |
| [43] | Wang R H, Zhang H, Zhang Y, et al. 2019. Development and evaluation of recombinase-aided amplification assays incorporating competitive internal controls for detection of human adenovirus serotypes 3 and 7. Virol J, 16(1): 86. |
| [44] | Wang S C, Zhuang Q Y, Jiang N, et al. 2023. Reverse transcription recombinase-aided amplification assay for avian influenza virus. Virus Genes, 59(3): 410-416. |
| [45] | Wang W J, Wang C G, Zhang Z C, et al. 2021. Recombinase-aided amplification-lateral flow dipstick assay: A specific and sensitive method for visual detection of avian infectious laryngotracheitis virus. Poult Sci, 100(3): 100895. |
| [46] | Wu T, Ge Y Y, Zhao K C, et al. 2020. A reverse-transcription recombinase-aided amplification assay for the rapid detection of N gene of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Virology, 549: 1-4. |
| [47] | Xu X M, Li Y, Xu Z Y, et al. 2022. TALE-induced immunity against the bacterial blight pathogen Xanthomonas oryzae pv. oryzae in rice. Phytopathol Res, 4(1): 47. |
| [48] | Yang Y, Zhou Y H, Sun J, et al. 2022. Research progress on cloning and function of Xa genes against rice bacterial blight. Front Plant Sci, 13: 847199. |
| [49] | Yu L, Yang C D, Ji Z J, et al. 2022a. First report of new bacterial leaf blight of rice caused by Pantoea ananatis in southeast China. Plant Dis, 106(1): 310. |
| [50] | Yu L, Yang C, Ji Z, et al. 2022b. Complete genomic data of Pantoea ananatis strain TZ39 associated with new bacterial blight of rice in China. Plant Dis, 106(2): 751-753. |
| [51] | Zhang C, Zhao T Y, Li J H, et al. 2024. Ultrasensitive and on-site diagnosis of rice bakanae disease based on CRISPR-LbCas12a coupled with LAMP. Pest Manag Sci, 80(12): 6527-6534. |
| [52] | Zhang H, Hu B S, Liu F Q. 2007. Double PCR technique for detection of pathogens causing rice bacterial leaf blight and streak. Plant Quarantine, 21(S1): 34-35. (in Chinese) |
| [53] | Zhang N, Li C, Dou X C, et al. 2022. Overview and future perspectives of microfluidic digital recombinase polymerase amplification (dRPA). Crit Rev Anal Chem, 52(8): 1969-1989. |
| [54] | Zhang R Q, Li G X, Li X N, et al. 2019. A rapid and sensitive recombinase aided amplification assay incorporating competitive internal control to detect Bordetella pertussis using the DNA obtained by boiling. Int J Infect Dis, 86: 108-113. |
| [55] | Zhu Z B, Li R, Zhang H W, et al. 2022. PAM-free loop-mediated isothermal amplification coupled with CRISPR/Cas12a cleavage (Cas-PfLAMP) for rapid detection of rice pathogens. Biosens Bioelectron, 204: 114076. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||