
Rice Science ›› 2019, Vol. 26 ›› Issue (2): 125-128.DOI: 10.1016/j.rsci.2018.07.002
• Letters • Previous Articles Next Articles
Hao Li, Ruiying Qin, Xiaoshuang Liu, Shengxiang Liao, Rongfang Xu, Jianbo Yang( ), Pengcheng Wei(
), Pengcheng Wei( )
)
Received:2018-06-09
Accepted:2018-07-26
Online:2019-03-04
Published:2018-12-18
Hao Li, Ruiying Qin, Xiaoshuang Liu, Shengxiang Liao, Rongfang Xu, Jianbo Yang, Pengcheng Wei. CRISPR/Cas9-Mediated Adenine Base Editing in Rice Genome[J]. Rice Science, 2019, 26(2): 125-128.
Add to citation manager EndNote|Ris|BibTeX
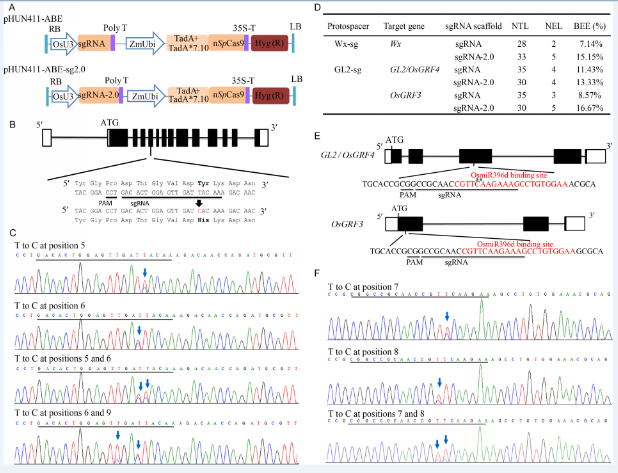
Fig. 1. Targeted adenine base editing (ABE) in rice genome.A, Schematic diagrams of rice codon-optimized tools pHUN411-ABE and pHUN411-ABE-sg2.0. B, Schematic illustration of the target site in the Wx gene. The protospacer adjacent motif (PAM) and sgRNA sequences are underlined. The desired point mutation of Wx-mq is indicated in red. C, Representative Sanger sequencing chromatograms of the ABE-edited Wx allele. Arrows mark the T to C substitutions. The protospacer sequence is underlined. D, The base editing efficiency of the different sgRNAs and different targets. E, Schematic illustration of the target site in the GL2 and OsGRF2 genes by the GL2-sg protospacer. The PAM and sgRNA sequences are underlined. The binding sequence of the miR396 is indicated in red. The asterisks show the mutation sites in the wild-type GL2 allele with large grain size. F, Representative Sanger sequencing chromatograms of the ABE-edited GL2 allele. RB, Right border; LB, Left border; NTL, Number of transgenic line; NEL, Number of edited line; BEE, Base editing efficiency.
| Element | Primer (5′-3′) | Function |
| ecTadA-XTEN-TadA*710 | F: TTACGCCAAGCTGCCCTTGGCGGCCGCATGAGCGAAGTGGA | ABE710 vector construction |
| R: GCGGGTCCTCCGGTGGTTCCGACAAGAAGTACTCCATCGG | ||
| nSpCas9(D10A) | F: GCGGGTCCTCCGGTGGTTCCGACAAGAAGTACTCCATCGGCCTCGCTATCGGCACCAAT | ABE710 vector construction |
| R: GCGAATTGAAGCTGCCCTTGGAGCTCTCACACCTTTCTTTTTTTTTTAGGAGAACCACCAGA | ||
| Wx-sg | F: GGCATTGTAATCAACTCCAGTGTC | sgRNA construction |
| R: AAACGACACTGGAGTTGATTACAA | ||
| GL2-sg | F: GGCATTCTTGAACGGTTGCGGCCG | sgRNA construction |
| R: AAACCGGCCGCAACCGTTCAAGAA | ||
| Wx | F: ACTGGTGATTTCAGGTTTGGGG | HRM |
| R: TGACCTGGCAAAGAAGGCTGA | ||
| F: GAGGTTTTTCCATTGCTACAAG | PCR sequencing | |
| R: GAAGTATGGGTTGTTGTTGAGG | ||
| GL2 | F: ACGGACGGCAAGAAATGGCG | HRM |
| R: CGCCGCAGAACCGACAACAG | ||
| F: CGGTTGCTACGATGTGCCTGTT | PCR sequencing | |
| R: AGAACCCAACGCCGAGCCAAAT | ||
| OsGRF3 | F: AAGAAGTGGCGGTGCTCCAAGG | HRM |
| R: GAGTGGTTCTGGAAGGCGGTGG | ||
| F: TGGCTGTAAAGCGTTCCATTCC | PCR sequencing | |
| R: GAGTATTAGACCCCAAGGCGAA | ||
| OsGRF5 | F: CCGCACCGACGGCAAGAAGT | HRM (off-target detection) |
| R: AGGGACAGACCGTGGGTTTTAG | ||
| F: GGATTCAATGTGCCTGTGCT | PCR sequencing (off target detection) | |
| R: GTCAATGAGATTAGAGGGGA |
Supplemental Table 1 The primers used in this study.
| Element | Primer (5′-3′) | Function |
| ecTadA-XTEN-TadA*710 | F: TTACGCCAAGCTGCCCTTGGCGGCCGCATGAGCGAAGTGGA | ABE710 vector construction |
| R: GCGGGTCCTCCGGTGGTTCCGACAAGAAGTACTCCATCGG | ||
| nSpCas9(D10A) | F: GCGGGTCCTCCGGTGGTTCCGACAAGAAGTACTCCATCGGCCTCGCTATCGGCACCAAT | ABE710 vector construction |
| R: GCGAATTGAAGCTGCCCTTGGAGCTCTCACACCTTTCTTTTTTTTTTAGGAGAACCACCAGA | ||
| Wx-sg | F: GGCATTGTAATCAACTCCAGTGTC | sgRNA construction |
| R: AAACGACACTGGAGTTGATTACAA | ||
| GL2-sg | F: GGCATTCTTGAACGGTTGCGGCCG | sgRNA construction |
| R: AAACCGGCCGCAACCGTTCAAGAA | ||
| Wx | F: ACTGGTGATTTCAGGTTTGGGG | HRM |
| R: TGACCTGGCAAAGAAGGCTGA | ||
| F: GAGGTTTTTCCATTGCTACAAG | PCR sequencing | |
| R: GAAGTATGGGTTGTTGTTGAGG | ||
| GL2 | F: ACGGACGGCAAGAAATGGCG | HRM |
| R: CGCCGCAGAACCGACAACAG | ||
| F: CGGTTGCTACGATGTGCCTGTT | PCR sequencing | |
| R: AGAACCCAACGCCGAGCCAAAT | ||
| OsGRF3 | F: AAGAAGTGGCGGTGCTCCAAGG | HRM |
| R: GAGTGGTTCTGGAAGGCGGTGG | ||
| F: TGGCTGTAAAGCGTTCCATTCC | PCR sequencing | |
| R: GAGTATTAGACCCCAAGGCGAA | ||
| OsGRF5 | F: CCGCACCGACGGCAAGAAGT | HRM (off-target detection) |
| R: AGGGACAGACCGTGGGTTTTAG | ||
| F: GGATTCAATGTGCCTGTGCT | PCR sequencing (off target detection) | |
| R: GTCAATGAGATTAGAGGGGA |
| Line | Target gene | Zygosity* | Genotype# |
| Wx-6 | Wx | Heterozygous | WT/T6toC |
| Wx-15 | Wx | Chimeric | WT/ T5toC/ T6toC |
| Wx-sg2.0-2 | Wx | Chimeric | WT/T5toC/T6toC |
| Wx-sg2.0-7 | Wx | Heterozygous | WT/T5toC |
| Wx-sg2.0-15 | Wx | Chimeric | WT/T6toC/T9toC |
| Wx-sg2.0-21 | Wx | Heterozygous | WT/T5T6toCC |
| Wx-sg2.0-22 | Wx | Heterozygous | WT/T6toC |
| *, The zygote of the mutant is putatively determined by the results of PCR sequencing and/or clone sequencing. #, WT, wild-type; TxtoC, the T at position x is mutated to C. | |||
Supplemental Table 2 Identification of targeted wx mutants induced by ABE tools in transgenic rice
| Line | Target gene | Zygosity* | Genotype# |
| Wx-6 | Wx | Heterozygous | WT/T6toC |
| Wx-15 | Wx | Chimeric | WT/ T5toC/ T6toC |
| Wx-sg2.0-2 | Wx | Chimeric | WT/T5toC/T6toC |
| Wx-sg2.0-7 | Wx | Heterozygous | WT/T5toC |
| Wx-sg2.0-15 | Wx | Chimeric | WT/T6toC/T9toC |
| Wx-sg2.0-21 | Wx | Heterozygous | WT/T5T6toCC |
| Wx-sg2.0-22 | Wx | Heterozygous | WT/T6toC |
| *, The zygote of the mutant is putatively determined by the results of PCR sequencing and/or clone sequencing. #, WT, wild-type; TxtoC, the T at position x is mutated to C. | |||
| Line | Target gene | Zygosity* | Genotype# |
| GL2-9 | GL2 | Heterozygous | WT/T7toC |
| OsGRF3 | Heterozygous | WT/T7toC | |
| GL2-17 | GL2 | Heterozygous | WT/T7toC |
| OsGRF3 | WT | WT | |
| GL2-22 | GL2 | Heterozygous | WT/T8toC |
| OsGRF3 | Heterozygous | WT/T7toC | |
| GL2-33 | GL2 | Heterozygous | WT/T7toC |
| OsGRF3 | Heterozygous | WT/T7toC | |
| GL2-sg2.0-6 | GL2 | Chimeric | WT/ T7toC/ T8toC |
| OsGRF3 | Heterozygous | WT/ T7toC | |
| GL2-sg2.0-11 | GL2 | WT | WT |
| OsGRF3 | Heterozygous | WT/ T7toC | |
| GL2-sg2.0-14 | GL2 | WT | WT |
| OsGRF3 | Chimeric | WT/ T7toC/ T7T8toCC | |
| GL2-sg2.0-23 | GL2 | Heterozygous | WT/ T8toC |
| OsGRF3 | Heterozygous | WT/ T7T8toCC | |
| GL2-sg2.0-30 | GL2 | Heterozygous | WT/ T8toC |
| OsGRF3 | Heterozygous | WT/ T7toC | |
| GL2-sg2.0-34 | GL2 | Heterozygous | WT/ T7toC |
| OsGRF3 | WT | WT | |
| *, The zygote of the mutant is putatively determined by the results of PCR-sequencing and/or clone-sequencing. #, WT, wild-type; TxtoC, the T at position x is mutated to C. | |||
Supplemental Table 3 Identification of targeted gl2 and/or osgrf3 mutants induced by ABE tools in transgenic rice.
| Line | Target gene | Zygosity* | Genotype# |
| GL2-9 | GL2 | Heterozygous | WT/T7toC |
| OsGRF3 | Heterozygous | WT/T7toC | |
| GL2-17 | GL2 | Heterozygous | WT/T7toC |
| OsGRF3 | WT | WT | |
| GL2-22 | GL2 | Heterozygous | WT/T8toC |
| OsGRF3 | Heterozygous | WT/T7toC | |
| GL2-33 | GL2 | Heterozygous | WT/T7toC |
| OsGRF3 | Heterozygous | WT/T7toC | |
| GL2-sg2.0-6 | GL2 | Chimeric | WT/ T7toC/ T8toC |
| OsGRF3 | Heterozygous | WT/ T7toC | |
| GL2-sg2.0-11 | GL2 | WT | WT |
| OsGRF3 | Heterozygous | WT/ T7toC | |
| GL2-sg2.0-14 | GL2 | WT | WT |
| OsGRF3 | Chimeric | WT/ T7toC/ T7T8toCC | |
| GL2-sg2.0-23 | GL2 | Heterozygous | WT/ T8toC |
| OsGRF3 | Heterozygous | WT/ T7T8toCC | |
| GL2-sg2.0-30 | GL2 | Heterozygous | WT/ T8toC |
| OsGRF3 | Heterozygous | WT/ T7toC | |
| GL2-sg2.0-34 | GL2 | Heterozygous | WT/ T7toC |
| OsGRF3 | WT | WT | |
| *, The zygote of the mutant is putatively determined by the results of PCR-sequencing and/or clone-sequencing. #, WT, wild-type; TxtoC, the T at position x is mutated to C. | |||
| Off-target locus | Transgenic plant | Number of detected plants* | Number of off-target mutants | |||
| HRM | Sequencing | HRM | Sequencing | |||
| OsGRF5 miR396 binding site | ABE-GL-sg | 35 | 24 | 0 | 0 | |
| ABE-sg2.0-GL-sg | 30 | 24 | 0 | 0 | ||
Supplemental Table 4 Detection of the putative off-target effects of the rice ABE tools on the high-similarity site of the GL2-sg protospacer.
| Off-target locus | Transgenic plant | Number of detected plants* | Number of off-target mutants | |||
| HRM | Sequencing | HRM | Sequencing | |||
| OsGRF5 miR396 binding site | ABE-GL-sg | 35 | 24 | 0 | 0 | |
| ABE-sg2.0-GL-sg | 30 | 24 | 0 | 0 | ||

Fig. 1. Sequence of rice codon-optimized ABE7.10.The sequence of wild type ecTadA, linker, TadA*7.10, nCas9(D10A) and NLS was labeled by blue, orange, red, black and cyan respectively.

Fig. 2 Genotyping of some targeted wx mutants.The mutations in the target region are indicated in red. The genotype of the wx targeted mutant was analyzed by PCR sequencing. The numbers located far right indicate the ratio of the number of clones with the corresponding genotype to the number of total sequenced clones. WT, wild-type; Tx to C, the T at position x is mutated to C.
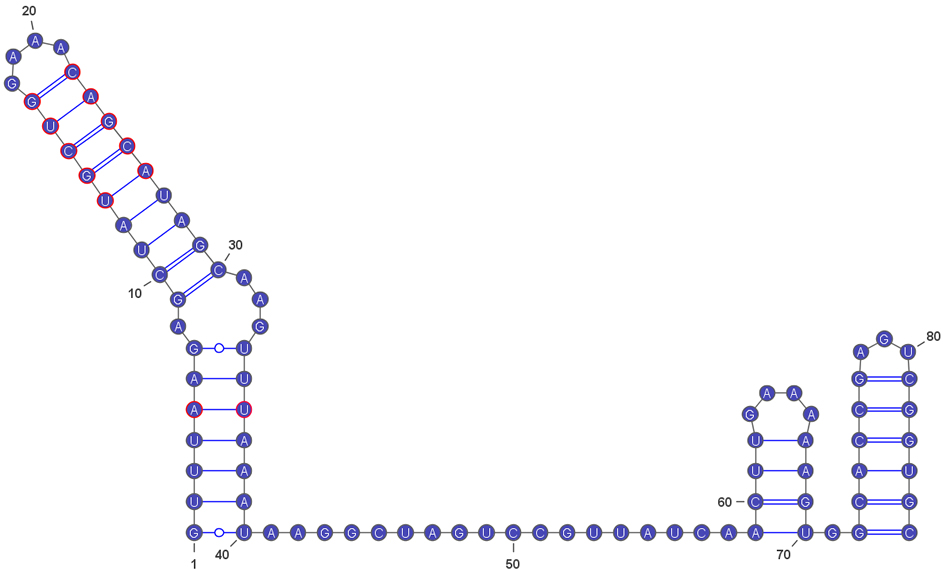
Fig. 3 The sequence of the extended sgRNA scaffold.The structures of the sgRNA-scaffold-2.0 sequence were generated with RNAfold and visualized using VARNA (Darty et al., 2009). The adapted and extended RNA is shown in red.
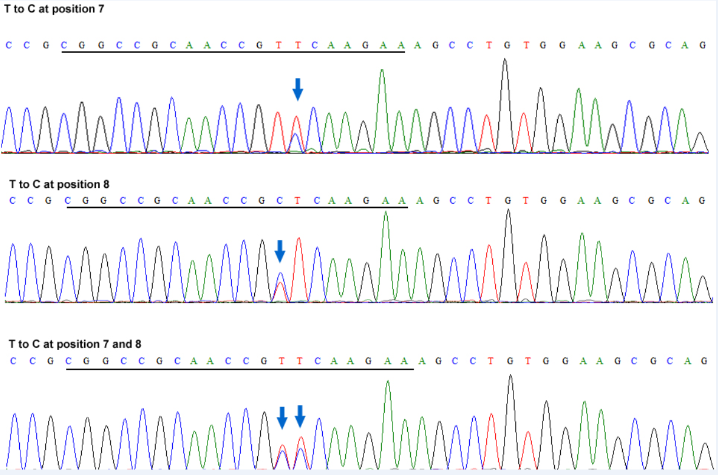
Fig. 4 Sanger sequencing chromatograms of the OsGRF3 target region of representative ABE-Gl2-sg edited lines.The targeted T to C mutation(s) is indicated by arrow(s). The target sequence is underlined.

Fig. 5 Genotyping of some transgenic plants targeted by ABE-GL2-sg.The mutations within the target region are indicated in red. The genotype of the GL2 and OsGRF3 target regions of the mutant was analyzed by PCR sequencing.
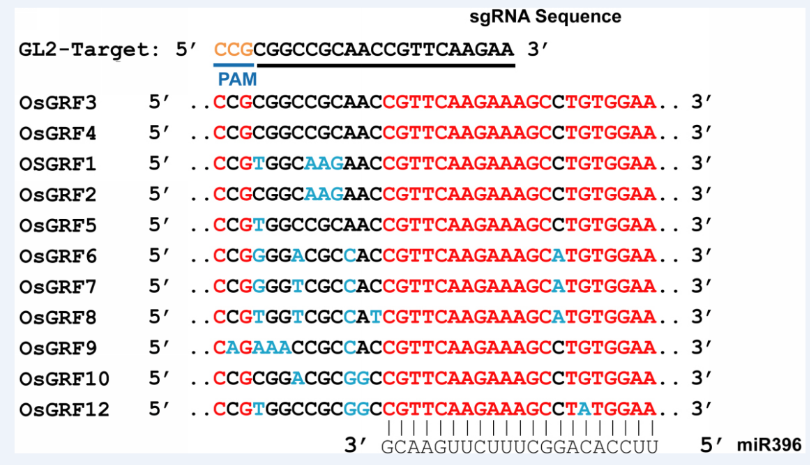
Fig. 6 Alignment of the miR396 binding region in the rice growth-regulating factor family.The GL2-sg shows 100% sequence identity with GL2 and OsGRF3, whereas there is one base mismatch with that of OsGRF5.

Fig. 7 The mutation of protein sequence resulting from the adenine base editor-induced editing.The amino acid of the target region of Wx (A) and GL2/OsGRF3 (B) was indicated. The PAM and protospacer sequence was labeled by blue and orange, respectively.
| [1] | Che R H, Tong H N, Shi B H, Liu Y Q, Fang S R, Liu D P, Xiao Y H, Hu B, Liu L C, Wang H R, Zhao M F, Chu C C.2015. Control of grain size and rice yield by GL2-mediated brassinosteroid responses.Nat Plants, 2: 15195. |
| [2] | Dang Y, Jia G X, Choi J, Ma H M, Anaya E, Ye C T, Shankar P, Wu H Q.2015. Optimizing sgRNA structure to improve CRISPR- Cas9 knockout efficiency.Genome Biol, 16: 280. |
| [3] | Gaudelli N M, Komor A C, Rees H A, Packer M S, Badran A H, Bryson D I, Liu D R.2017. Programmable base editing of A∙T to G∙C in genomic DNA without DNA cleavage.Nature, 551: 464-471. |
| [4] | Hu X X, Meng X B, Liu Q, Li J Y, Wang K J.2018. Increasing the efficiency of CRISPR-Cas9-VQR precise genome editing in rice.Plant Biotechnol J, 16(1): 292-297. |
| [5] | Hua K, Tao X P, Yuan F T, Wang D, Zhu J K.2018. Precise A∙T to G∙C base editing in the rice genome.Mol Plant, 11(4): 627-630. |
| [6] | Koblan L W, Doman J L, Wilson C, Levy J M, Tay T, Newby G A, Maianti J P, Raguram A, Liu D R.2018. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction.Nat Biotechnol, doi: 10.1038/nbt.4172. |
| [7] | Komor A C, Kim Y B, Packer M S, Zuris J A, Liu D R.2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage.Nature, 533: 420-424. |
| [8] | Li C, Zong Y, Wang Y P, Jin S, Zhang D B, Song Q N, Zhang R, Gao C X.2018. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion.Genome Biol, 19: 59. |
| [9] | Liu H H, Guo S Y, Xu Y Y, Li C H, Zhang Z Y, Zhang D J, Xu S J, Zhang C, Chong K.2014. OsmiR396d-regulatedOsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol, 165(1): 160-174. |
| [10] | Lu Y, Zhu J K.2017. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system.Mol Plant, 10(3): 523-525. |
| [11] | Miki D, Zhang W X, Zeng W J, Feng Z Y, Zhu J K.2018. CRISPR/ Cas9-mediated gene targeting inArabidopsis using sequential transformation. Nat Commun, 9: 1967. |
| [12] | Sato H, Suzuki Y, Sakai M, Imbe T.2002. Molecular characterization ofWx-mq, a novel mutant gene for low-amylose content in endosperm of rice(Oryza sativa L.). Jpn J Breeding, 52(2): 131-135 |
| [13] | Xing H L, Dong L, Wang Z P, Zhang H Y, Han C Y, Liu B, Wang X C, Chen Q J.2014. A CRISPR/Cas9 toolkit for multiplex genome editing in plants.BMC Plant Biol, 14: 327. |
| [14] | Xu R F, Yang Y C, Qin R Y, Li H, Qiu C H, Li L, Wei P C, Yang J B.2016. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice.J Genet Genom, 43(8): 529-532. |
| [15] | Yan F, Kuang Y J, Ren B, Wang J W, Zhang D W, Lin H H, Yang B, Zhou X P, Zhou H B.2018. Highly efficient A∙T to G∙C base editing by Cas9n-guided tRNA adenosine deaminase in rice.Mol Plant, 11(4): 631-634. |
| [16] | Zong Y, Wang Y P, Li C, Zhang R, Chen K L, Ran Y D, Qiu J L, Wang D W, Gao C.2017. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion.Nat Biotechnol, 35(5): 438-440. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||