
Rice Science ›› 2020, Vol. 27 ›› Issue (3): 180-183.DOI: 10.1016/j.rsci.2020.04.002
• Letter • Previous Articles Next Articles
Sanfeng Li1, Lan Shen1, Ping Hu1, Xianmei Wu1, Qiaoling Yuan2, Yuchun Rao3( ), Qian Qian1, Kejian Wang1, Xudong Zhu1, Lianguang Shang2(
), Qian Qian1, Kejian Wang1, Xudong Zhu1, Lianguang Shang2( ), Yuexing Wang1(
), Yuexing Wang1( )
)
Received:2019-07-11
Accepted:2019-10-01
Online:2020-05-28
Published:2020-01-17
About author:# These authors contributed equally to this work
Sanfeng Li, Lan Shen, Ping Hu, Xianmei Wu, Qiaoling Yuan, Yuchun Rao, Qian Qian, Kejian Wang, Xudong Zhu, Lianguang Shang, Yuexing Wang. A Method for Effectively Overcoming Tight Functional Linkage Between Genes in Rice by CRISPR/Cas9 System[J]. Rice Science, 2020, 27(3): 180-183.
Add to citation manager EndNote|Ris|BibTeX
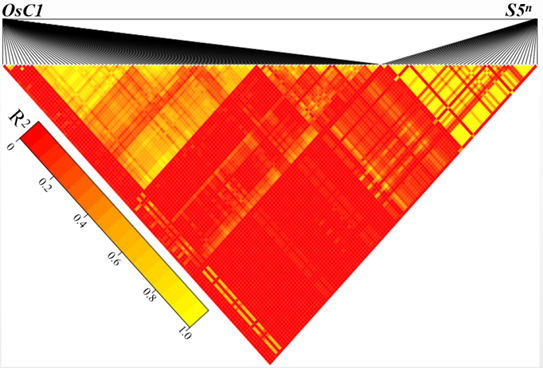
Supplemental Fig. 1. Pairwise linkage disequilibrium heatmap (LD heatmap) of the OsC1 and S5n gene.Heatmap displaying pairwise linkage disequilibrium between polymorphic single nucleotide polymorphisms (SNPs) of OsC1 and S5n gene. The heat color scale represents squared correlation (R2) between pairs of SNPs. Color intensity corresponds to higher R2 values according to the legend. The higher the R2 value between the SNPs, the tighter the linkage.
| Plant line was sequenced | Target | Gene | Off-target site | No. of mismatching bases | No. of plants sequenced | No. of plants with mutations | Mutation rate (%) | |
|---|---|---|---|---|---|---|---|---|
| osc1-2 | OsC1 | Os06g0205100 | GAGCCAATGGCCTTCGCCATGGG | 3 | 36 | 0 | 0 | |
| CTCCCTCTTGCCTTCGCCATGGG | 4 | 36 | 0 | 0 | ||||
| GCGCCGCTGGCGTTCGCCATCGG | 3 | 36 | 0 | 0 | ||||
| CCGCCCATGGCCTTCGCCATAGG | 4 | 36 | 0 | 0 | ||||
Supplemental Table 1. Mutations detected in putative CRISPR/Cas9 off-target sites.
| Plant line was sequenced | Target | Gene | Off-target site | No. of mismatching bases | No. of plants sequenced | No. of plants with mutations | Mutation rate (%) | |
|---|---|---|---|---|---|---|---|---|
| osc1-2 | OsC1 | Os06g0205100 | GAGCCAATGGCCTTCGCCATGGG | 3 | 36 | 0 | 0 | |
| CTCCCTCTTGCCTTCGCCATGGG | 4 | 36 | 0 | 0 | ||||
| GCGCCGCTGGCGTTCGCCATCGG | 3 | 36 | 0 | 0 | ||||
| CCGCCCATGGCCTTCGCCATAGG | 4 | 36 | 0 | 0 | ||||
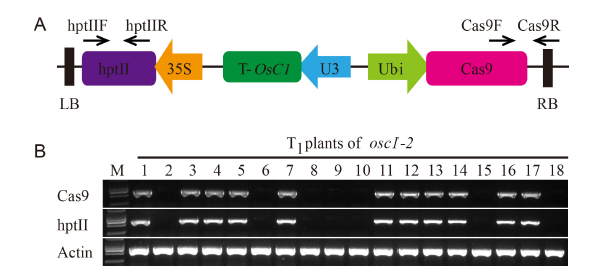
Supplemental Fig. 2. Isolation of possible transgene-free T1 plants from osc1-2.A, A schematic diagram of the position of specific primers used to amplify the hptII and Cas9 sequences in T1 plants. B, hptII and Cas9 DNA fragments could not be detected in some T1 plants of the osc1-2 mutant (numbers 2, 6, 8, 9, 10, 15, and 18). The endogenous OsACTIN1 was used as a control. M, DNA molecular weight marker.
| Primer name | Sequence (5′-3′) | Purpose |
|---|---|---|
| OsC1-g++ OsC1-g-- T3 OsC1-JC-F OsC1-JC-R hptII F | GGCAGCGCCACTTGCCTTCGCCAT AAACATGGCGAAGGCAAGTGGCGC ATTAACCCTCACTAAAGGGA CCAGATCGCTCAGTCTCACA TAGGCCGGAGATAGTTGAGC GCTGTTATGCGGCCATTGTC | Vector construction Vector construction Vector construction Detection of target mutations Detection of target mutations Genotyping |
| hptII R | GACGTCTGTCGAGAAGTTTC | Genotyping |
| Cas9 F | ACCAGACACGAGACGACTAA | Genotyping |
| Cas9 R | ATCGGTGCGGGCCTCTTC | Genotyping |
| Actin-F | TGCTGACAGGATGAGCAAGG | Genotyping |
| Actin-R | CCCAACCATGCAAAGCTCAC | Genotyping |
| OsC1-qPCR-F | ATGGGGAGGAGAGCTTGCTG | qPCR |
| OsC1-qPCR-R | CGGAGATAGTTGAGCCACC | qPCR |
| Ubi-qPCR-F | GCTCCGTGGCGGTATCAT | qPCR |
| Ubi-qPCR-R | CGGCAGTTGACAGCCCTAG | qPCR |
| S5n-JD-F | CTTCATTCCCAGCGAGCGG | Genotyping |
| S5n-JD-R | ATGGGCGGAGGCATTGGT | Genotyping |
| OsC1-JD-F | GCAAAGGAAGGGATGAAGAG | Genotyping |
| OsC1-JD-R | CGTCATCGCCGTCTCCTAATT | Genotyping |
Supplemental Table 2. Primers used in this research.
| Primer name | Sequence (5′-3′) | Purpose |
|---|---|---|
| OsC1-g++ OsC1-g-- T3 OsC1-JC-F OsC1-JC-R hptII F | GGCAGCGCCACTTGCCTTCGCCAT AAACATGGCGAAGGCAAGTGGCGC ATTAACCCTCACTAAAGGGA CCAGATCGCTCAGTCTCACA TAGGCCGGAGATAGTTGAGC GCTGTTATGCGGCCATTGTC | Vector construction Vector construction Vector construction Detection of target mutations Detection of target mutations Genotyping |
| hptII R | GACGTCTGTCGAGAAGTTTC | Genotyping |
| Cas9 F | ACCAGACACGAGACGACTAA | Genotyping |
| Cas9 R | ATCGGTGCGGGCCTCTTC | Genotyping |
| Actin-F | TGCTGACAGGATGAGCAAGG | Genotyping |
| Actin-R | CCCAACCATGCAAAGCTCAC | Genotyping |
| OsC1-qPCR-F | ATGGGGAGGAGAGCTTGCTG | qPCR |
| OsC1-qPCR-R | CGGAGATAGTTGAGCCACC | qPCR |
| Ubi-qPCR-F | GCTCCGTGGCGGTATCAT | qPCR |
| Ubi-qPCR-R | CGGCAGTTGACAGCCCTAG | qPCR |
| S5n-JD-F | CTTCATTCCCAGCGAGCGG | Genotyping |
| S5n-JD-R | ATGGGCGGAGGCATTGGT | Genotyping |
| OsC1-JD-F | GCAAAGGAAGGGATGAAGAG | Genotyping |
| OsC1-JD-R | CGTCATCGCCGTCTCCTAATT | Genotyping |
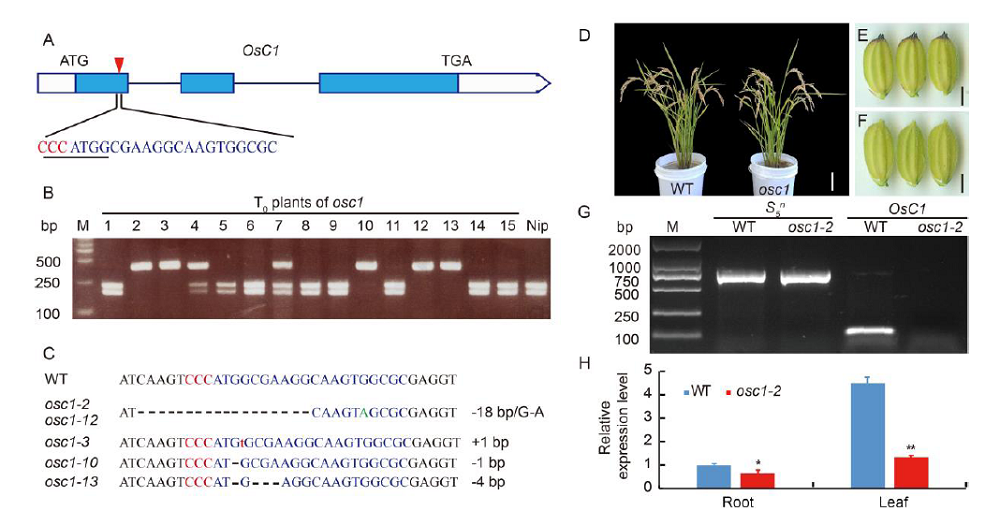
Fig. 1. CRISPR/Cas9 induced mutation in OsC1 gene break the tight functional linkage between OsC1 and S5n.A, Schematic representation of OsC1 gene structure and the genomic RNA (gRNA) target site on its genomic sequence. Introns are depicted as lines, and exons are depicted as blue boxes. The targeted site is labeled with a red arrowhead, and its sequence is depicted by blue uppercase letters; the protospacer adjacent motif (PAM) (CCC) is marked red. The recognition sequence of NcoI is underlined. B, Identification of mutations in osc1 via PCR-based restriction enzyme assay in the T0 generation. The PCR products of osc1-2, -3, -10, -12 and -13 are resistant to NcoI digestion. Nipponbare (Nip) was used as the negative control. M, Marker. C, Sequencing results at the target sites in T0 plants. The targeted sequence is highlighted in blue, and the PAM sequence is in red. Green uppercase letter indicates substituted nucleotide. Red lowercase letter indicates inserted nucleotide. Black dashes indicate deleted nucleotides. ‘+’ and ‘-’ on the right indicate insertion and deletion, respectively, and numbers indicate number of nucleotides. WT, Wild type. D, Phenotypes of WT and osc1-2 plants. E, Grain shape of WT. F, Grain shape of osc1-2. G, Analysis of PCR production of OsC1 and S5n in WT and osc1-2 on 1% agarose gel. M, Marker. H, Quantitative real-time PCR analysis of the transcription of OsC1 in roots and leaves of WT and osc1-2 plants. *, P < 0.05; **, P < 0.01 (Student’s t-test).
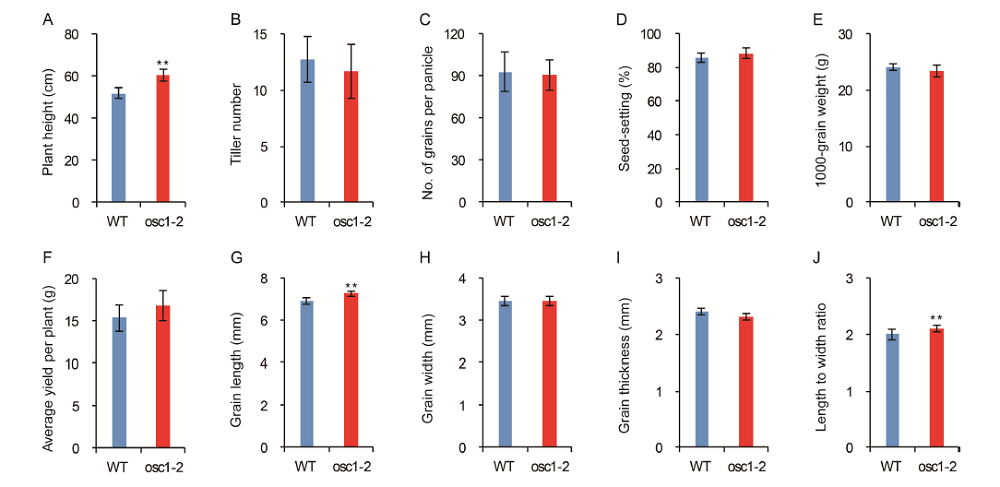
Supplemental Fig. 3. Characterization of agronomic traits of wild-type (WT) and osc1-2 plants.A, Plant height. B, Tiller numbers per plant. C, Grain number per panicle. D, Seed-setting. E, Weight of 1,000 grains. F, Average yield per plant. G, Grain length. H, Grain width. I, Grain thickness. J, Length-to-width ratio. All data are from mature plants grown in the field under normal agricultural conditions. Data are presented as Mean ± SD (n = 10). Differences between the two genotypes were compared using Student’s t-test (**, P < 0.01).
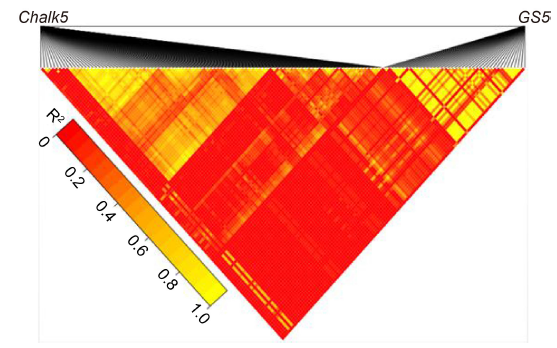
Supplemental Fig. 4. Pairwise linkage disequilibrium heatmap (LD heatmap) in the Chalk5 and GS5 genes.Heatmap displaying pairwise linkage disequilibrium between polymorphic SNPs of Chalk5 and GS5 genes. The heat color scale represents R2 between pairs of SNPs. Color intensity corresponds to higher R2 values according to the legend. The higher the R2 value between SNPs, the tighter the linkage.
| [1] | Aizza L C B, Dornelas M C. 2011. A genomic approach to study anthocyanin synthesis and flower pigmentation in passionflowers.J Nucl Acids, 2011: 371517. |
| [2] | Baltes N J, Voytas D F.2015. Enabling plant synthetic biology through genome engineering.Trends Biotechnol, 33(2): 120-131. |
| [3] | Fan F J, Fan Y Y, Du J H, Zhuang J Y.2007. Fine mapping of C(chromogen for anthocyanin) gene in rice. Chin J Rice Sci, 21(5): 454-458. (in Chinese with English abstract) |
| [4] | Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M.2009. Loss of function of a proline-containing protein confers durable disease resistance in rice.Science, 325: 998-1001. |
| [5] | Hou F Y, Wang Q M, Li A X.2009. Study progress on anthocyanidin synthase of plants.Chin Agric Sci Bull, 25(21): 188-190. |
| [6] | Huang S W, Weigel D, Beachy R N, Li J Y.2016. A proposed regulatory framework for genome-edited crops.Nat Genet, 48(2): 109-111. |
| [7] | Li M R, Li X X, Zhou Z J, Wu P Z, Fang M C, Pan X P, Lin Q P, Luo W B, Wu G J, Li H Q.2016. Reassessment of the four yield-related genesGn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci, 7: 377. |
| [8] | Li Y B.2011. Cloning and functional analysis of a major QTL, GS5 for grain size/weight and Chalk5 for chalkiness rate in rice. [PhD Thesis]. Wuhan, China: Huazhong Agricultural University. |
| [9] | Lin T Z, Sun L T, Gong H B, Wang Y H, Liu L L, Zhao Z G, Jiang L, Wan J M.2019. Identification and gene mapping of awhite-stripe leaf after transplanting at low temperature mutant in rice. Chin J Rice Sci, 33(1): 1-11. |
| [10] | Luo X, Ji S D, Yuan P R, Lee H S, Kim D M, Balkunde S, Kang J W, Ahn S.2013. QTL mapping reveals a tight linkage between QTLs for grain weight and panicle spikelet number in rice.Rice, 6(1): 33. |
| [11] | Pang A Y, Jin L, Chen J Y, Liu C L, Ruan Y.2018. Obtain of double mutant of closely linked genesM320 and M330 by CRISPR/Cas9 system. Mol Plant Breeding, 16(13): 4301-4307. |
| [12] | Reddy A R.1996. Genetic and molecular analysis of the anthocyanin pigmentation pathway in rice.Plant Mol Biol, 32(4): 735-743. |
| [13] | Saitoh K, Onishi K, Mikami I, Thidar K, Sano Y.2004. Allelic diversification at theC (OsC1) locus of wild and cultivated rice: Nucleotide changes associated with phenotypes. Genetics, 168(2): 997-1007. |
| [14] | Sakamoto W, Ohmori T, Kageyama K, Miyazaki C, Saito A, Murata M, Noda K, Maekawa M.2001. ThePurple leaf (Pl) locus of rice: The Pl(w) allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol, 42: 982-991. |
| [15] | Shao G N, Xie L H, Jiao G A, Wei X J, Sheng Z H, Tang S Q, Hu P S.2017. CRISPR/CAS9-mediated editing of the fragrant geneBadh2 in rice. Chin J Rice Sci, 31(2): 216-222. (in Chinese with English abstract) |
| [16] | Sheng S L, Yu B, Li C, Lu J F.2008. Study on the location of chromogen gene and its physical distance from broad-affinity genes.Acta Agric Jiangxi, 20(9): 27-28. (in Chinese with English abstract) |
| [17] | Sun Y W, Jiao G A, Liu Z P, Zhang X, Li J Y, Guo X P, Du W M, Du J L, Francis F, Zhao Y D, Xia L Q.2017. Generation of high- amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes.Front Plant Sci, 8: 298. |
| [18] | Tang L, Mao B G, Li Y K, Lv Q M, Zhang L P, Chen C Y, He H J, Wang W P, Zeng X F, Shao Y, Pan Y L, Hu Y Y, Peng Y, Fu X Q, Li H Q, Xia S T, Zhao B R.2017. Knockout ofOsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci Rep, 7: 14438. |
| [19] | Zhang J S, Zhang H, Botella J R, Zhu J K.2018. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of theWaxy gene in elite rice varieties. J Integr Plant Biol, 60(5): 369-375. |
| [20] | Zhao S S, Wang C H, Ma J, Wang S, Tian P, Wang J L, Cheng Z J, Zhang X, Guo X P, Lei C L.2016. Map-based cloning and functional analysis of the chromogen geneC in rice(Oryza sativa L.). J Plant Biol, 59(5): 496-505. |
| [21] | Zhou H, He M, Li J, Chen L, Huang Z F, Zheng S Y, Zhu L Y, Ni E D, Jiang D G, Zhao B R, Zhuang C X.2016. Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediatedTMS5 editing system. Sci Rep, 6: 37395. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||