
Rice Science ›› 2021, Vol. 28 ›› Issue (1): 1-5.DOI: 10.1016/j.rsci.2020.11.001
• Letter • Next Articles
Huimei Wang1, Yuxuan Hou1, Shuang Wang1,2, Xiaohong Tong1, Liqun Tang1, Adijat Ajadi Abolore1, Jian Zhang1( ), Yifeng Wang1(
), Yifeng Wang1( )
)
Received:2019-12-20
Accepted:2020-05-30
Online:2021-01-28
Published:2021-01-28
Huimei Wang, Yuxuan Hou, Shuang Wang, Xiaohong Tong, Liqun Tang, Adijat Ajadi Abolore, Jian Zhang, Yifeng Wang. WRKY72 Negatively Regulates Seed Germination Through Interfering Gibberellin Pathway in Rice[J]. Rice Science, 2021, 28(1): 1-5.
Add to citation manager EndNote|Ris|BibTeX
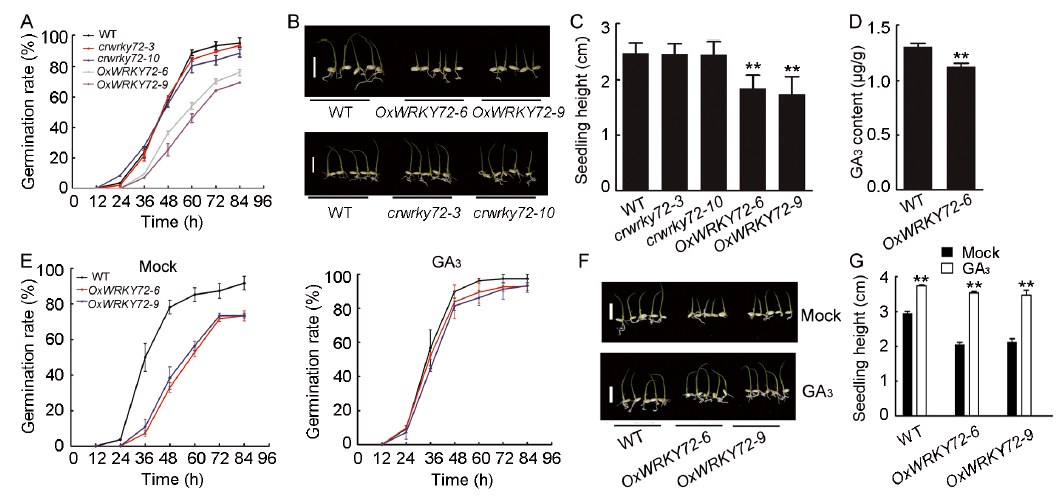
Fig. 1. Seed germination characteristics of overexpression lines OxWRKY72 and mutant lines crwrky72.A, Germination time courses of the wild type (WT), overexpression lines OxWRKY72 and mutant lines crwrky72, respectively. B, Germination phenotypes of the WT, OxWRKY72 and crwrky72 grown on 1/2 Murashige and Skoog (MS) medium for 4 d. Scale bars, 1 cm. C, Seedling heights of the WT, OxWRKY72 and crwrky72 in accordance to B. D, GA3 content in the germinating embryos of the WT and OxWRKY72-6. E, Germination time courses of the WT and OxWRKY72 under mock or 1.5 µmol/L GA3 treatment. F, Germination phenotypes of the WT and OxWRKY72 under mock or GA3 treatment for 4 d. Scale bars, 2 cm. G, Seedling heights of the WT and OxWRKY72 lines in accordance to F. Error bars indicate SD with triple biological replicates (each replicate containing 50 seeds) in A and E, 50 biological replicates in C and G, and triple biological replicates in D. Asterisks indicate the significant differences between the WT and transgenic lines as determined by the Student’s t-test analysis. **, P < 0.01.
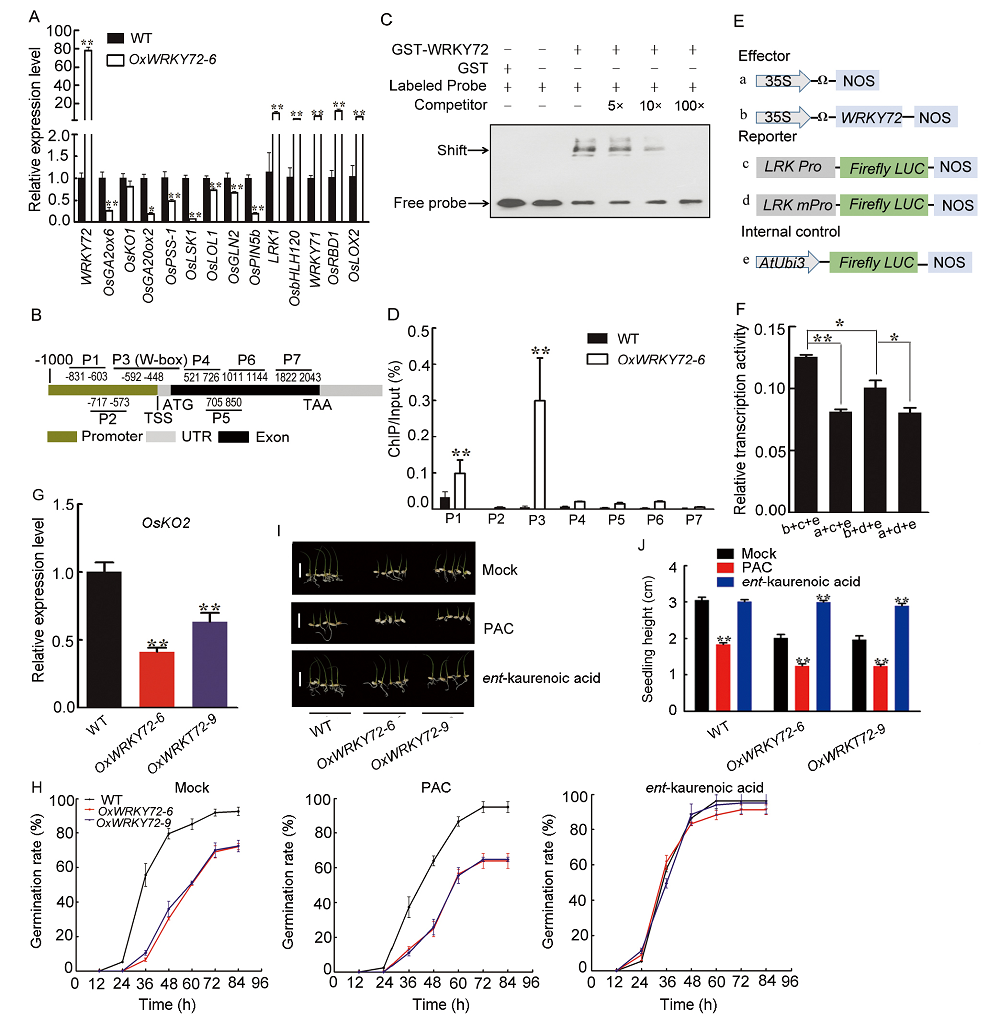
Fig. 2. WRKY72 mediates seed germination by WRKY72-LRK1-OsKO2 pathway. A, Real-time PCR (qRT-PCR) validation of the differentially expressed genes (DEGs) revealed by RNA-sequencing (RNA-seq) experiments. cDNA of germinating embryos grown on 1/2 Murashige and Skoog (MS) medium for 2 d was used as templates. B, Probe positions on LRK1 promoter and genome. Grey, black and yellow boxes represent untranslational regions, coding sequence and promoter regions, respectively. Transcription starting site (TSS) was set as 0. Numbers indicate the distances (bps) to the TSS. C, Electrophoretic mobility shift assay (EMSA) to show GST-WRKY72 specifically binds with the probe 3 (P3) region on the promoter of LRK1 in B. Purified GST, GST-bZIP72 was detected with anti-GST antibody. The 5-, 10- and 100-fold excess non-labeled probes were applied for competition. D, Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assay to show WRKY72 binding to the promoter regions of LRK1. P1?P7 represent the regions shown in B detected by ChIP-qPCR, respectively. The enrichment values were normalized to the Input. IgG immunoprecipitated DNA was used as a control. E and F, Luciferase (LUC) transient transcriptional activity assay in rice protoplast. mPro, The promoter of LRK1 with G-box mutated. G, qRT-PCR analysis for the transcript accumulation of OsKO2 in OxWRKY72 germinating embryos grown on half-strength MS medium for 2 d. H, Germination time courses of the wild type (WT) and OxWRKY72 under mock, 3 µmol/L ent-kaurenoic acid or 10 µmol/L paclobutrazol (PAC) treatments, respectively. I, Germination phenotypes of the WT and OxWRKY72 under mock, ent-kaurenoic acid or PAC treatments for 4 d. Scale bars, 2 cm. J, Seedling heights of the WT and OxWRKY72 in accordance to I. Data represent Mean ± SD (n = 3) in A, D, F and G, n = 3 (each replicate containing 50 seeds) in H, and n = 50 in J. Asterisks indicate the significant differences as determined by the Student’s t-test analysis (*, P < 0.05; **, P < 0.01).
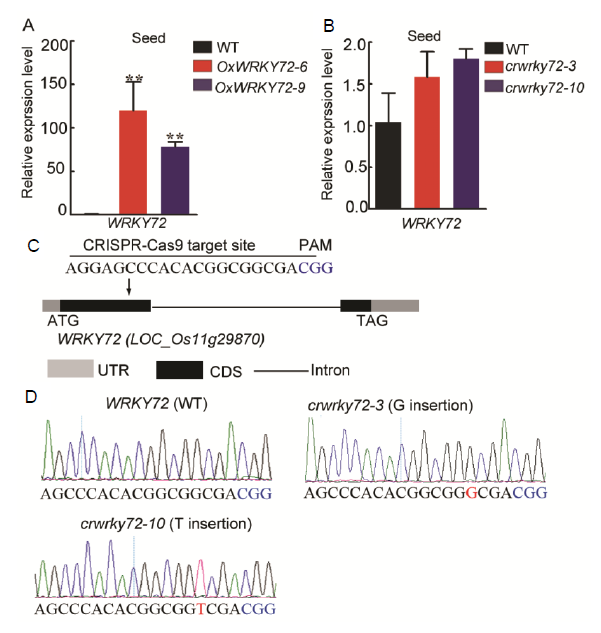
Fig. S1. Molecular characterization of OxWRKY72 and crwrky72 mutants. A and B, qRT-PCR analysis for the transcript accumulation of WRKY72 in OxWRKY72 (A) and crwrky72 (B) germinating embryos grown on half-strength MS medium for 2 d. Error bars indicate SD with biological triplicates (n = 3). Asterisks indicate the significance of differences between the WT and transgenic lines as determined by the Student’s t-test analysis, ** P < 0.01. WT, Wild type; MS, Murashige and Skoog medium. C, Schematic presentation of the gene structure of WRKY72 and CRISPR-cas9 editing site. Grey boxes represent untranslational regions (UTR), black boxes indicate exons (CDS), and black line refers to intron. PAM, Protospacer adjacent motif. D, Sanger sequencing chromatograph of the CRISPR-cas9 target site in homozygous mutants of crwrky72. The letters in red represent the mutant sites. The letters in blue represent the PAM sequence.

Fig. S2. Occurrence of cis-regulatory elements in promoters of OsGA2ox6 and LRK1.Black boxes represent genomics regions, and white boxes represent promoters. Transcription starting site (TSS) is set as 0. Numbers indicate the distances (bps) to the TSS.
| [1] | Eulgem T, Rushton P J, Robatzek S, Somssich I E. 2000. The WRKY superfamily of plant transcription factors. Trends Plant Sci, 5(5): 199-206. |
| [2] | Hou Y X, Wang Y F, Tang L Q, Tong X H, Wang L, Liu L M, Huang S W, Zhang J. 2019. SAPK10-mediated phosphorylation on WRKY72 releases its suppression on jasmonic acid biosynthesis and bacterial blight resistance. iScience, 16: 499-510. |
| [3] | Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M. 2004. A rice semi-dwarf gene,Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol Biol, 54(4): 533-547. |
| [4] | Kaneko M, Itoh H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M. 2002. The alpha-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol, 128(4): 1264-1270. |
| [5] | Peng X X, Hu Y J, Tang X K, Zhou P L, Deng X B, Wang H H, Guo Z J. 2012. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta, 236(5): 1485-1498. |
| [6] | Qiu D Y, Xiao J, Ding X H, Xiong M, Cai M, Cao Y L, Li X H, Xu C G, Wang S P. 2007. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate- dependent signaling. Mol Plant Microbe Interact, 20(5): 492-499. |
| [7] | Ramamoorthy R, Jiang S Y, Kumar N, Venkatesh P N, Ramachandran S. 2008. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol, 49(6): 865-879. |
| [8] | Reinecke D M, Wickramarathna A D, Ozga J A, Kurepin L V, Jin A L, Good A G, Pharis R P. 2013. Gibberellin 3-oxidase gene expression patterns influence gibberellin biosynthesis, growth, and development in pea. Plant Physiol, 163(2): 929-945. |
| [9] | Rushton P J, Macdonald H, Huttly A K, Lazarus C M, Hooley R. 1995. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol Biol, 29(4): 691-702. |
| [10] | Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal G K, Takeda S, Abe K, Miyao A, Hirochika H, Kitano H, Ashikari M, Matsuoka M. 2004. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol, 134(4): 1642-1653. |
| [11] | Song Y, Chen L G, Zhang L P, Yu D Q. 2010. Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J Biosci, 35(3): 459-471. |
| [12] | Swain S M, Singh D P, Helliwell C A, Poole A T. 2005. Plants with increased expression of ent-kaurene oxidase are resistant to chemical inhibitors of this gibberellin biosynthesis enzyme. Plant Cell Physiol, 46(2): 284-291. |
| [13] | Ulker B, Somssich I E. 2004. WRKY transcription factors: From DNA binding towards biological function. Curr Opin Plant Biol, 7(5): 491-498. |
| [14] | Wang H H, Meng J, Peng X X, Tang X K, Zhou P L, Xiang J H, Deng X B. 2015. Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol Biol, 89(1/2): 157-171. |
| [15] | Wu J H, Zhu C F, Pang J H, Zhang X R, Yang C L, Xia G X, Tian Y C, He C Z. 2014. OsLOL1, a C2C2-type zinc finger protein, interacts with OsbZIP58 to promote seed germination through the modulation of gibberellin biosynthesis in Oryza sativa. Plant J, 80(6): 1118-1130. |
| [16] | Xie Z, Zhang Z L, Zou X L, Huang J, Ruas P, Thompson D, Shen Q J. 2005. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol, 137(1): 176-189. |
| [17] | Xie Z, Zhang Z L, Zou X L, Yang G X, Komatsu S, Shen Q J. 2006. Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J, 46(2): 231-242. |
| [18] | Yang M F, Qi W W, Sun F, Zha X J, Chen M L, Huang Y Q, Feng Y Q, Yang J S, Luo X J. 2013. Overexpression of rice LRK1 restricts internode elongation by down-regulating OsKO2. Biotechnol Lett, 35(1): 121-128. |
| [19] | Ye H, Feng J H, Zhang L H, Zhang J F, Mispan M S, Cao Z Q, Beighley D H, Yang J C, Gu X Y. 2015. Map-based cloning of seed dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol, 169(3): 2152-2165. |
| [20] | Zentella R, Zhang Z L, Park M, Thomas S G, Endo A, Murase K, Fleet C M, Jikumaru Y, Nambara E, Kamiya Y, Sun T P. 2007. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell, 19(10): 3037-3057. |
| [21] | Zhang Z L, Xie Z, Zou X L, Casaretto J, Ho T H, Shen Q J. 2004. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol, 134(4): 1500-1513. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||