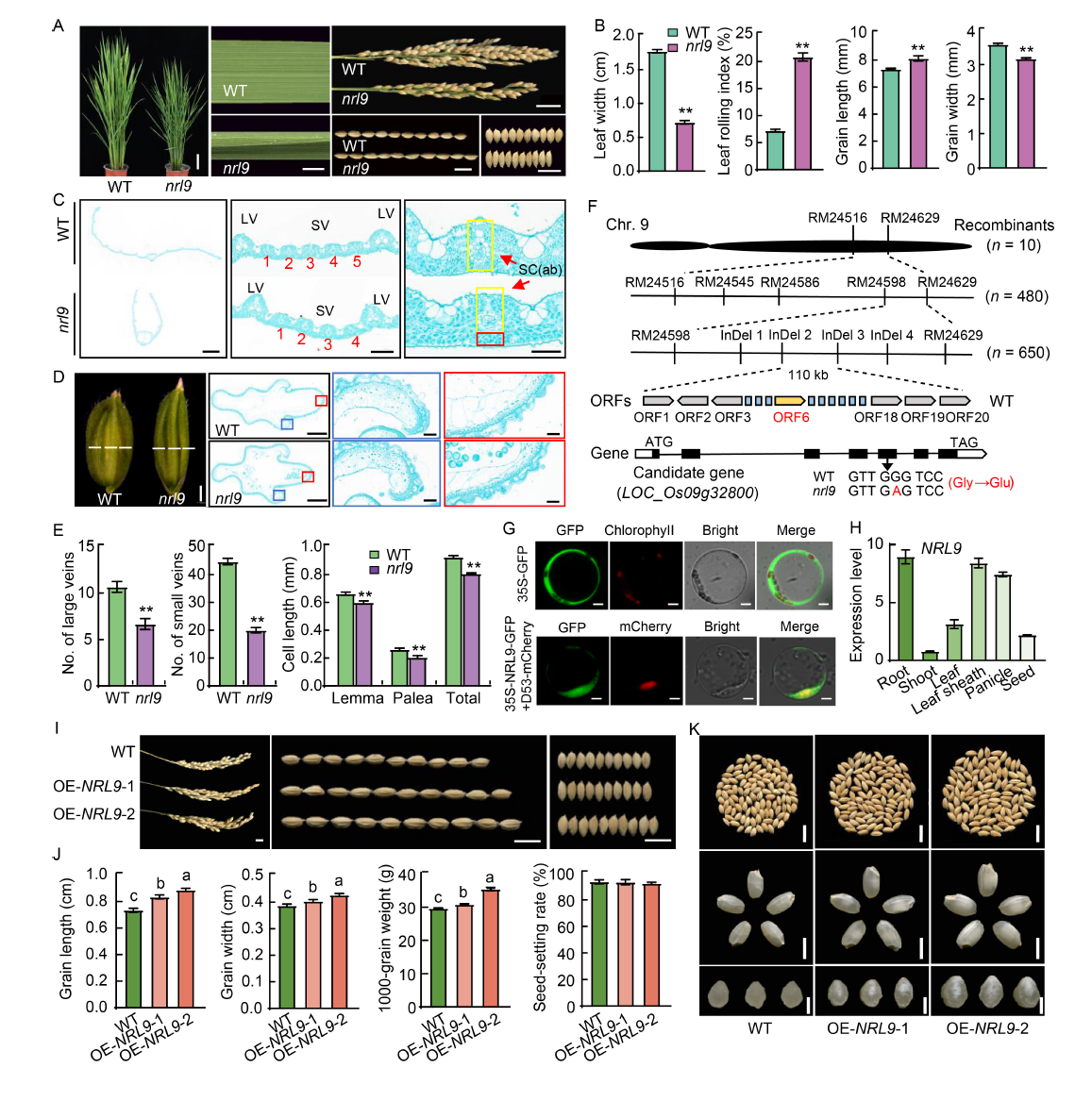
Fig. 1. NARROW AND ROLLED LEAF 9 (NRL9) is a novel gene regulating leaf morphology and grain size in rice. A, Plant and leaf morphologies at the heading stage, and panicle and grain morphologies at the maturity stage of wild type (WT) and nrl9 mutant. Scale bars are 10, 1, 2, 1, and 1 cm from left to right, respectively. B, Leaf width, leaf rolling index, grain length, and grain width for WT and nrl9 mutants. C, Cytological analysis of leaf cross-sections for WT and nrl9 mutants. LV, Large vein; SV, Small vein; SC(ab), Sclerenchyma cells (abaxial). The yellow and red boxes indicate the normal development of sclerenchyma cells and the partially developed defective sclerenchyma cells, respectively. The numbers in the figure indicate the sequential count of SVs between two LVs. Scale bars are 1000, 100, and 50 μm from left to right, respectively. D, Morphology of non-flowered spikelet hulls. The red and blue boxes indicate the local enlarged views of the lemma and palea surfaces of the glume wax sections, respectively. Scale bars are 1000, 500, 50, and 50 μm from left to right, respectively. E, Number of large and small veins in WT and nrl9 mutant leaves, as well as cell lengths of lemma, palea, and total. F, Map-based cloning of NRL9 gene. NRL9 was mapped to a 110-kb interval on chromosome 9. Sequencing revealed a non-synonymous mutation from G to A in exon of the 6th open reading frame (ORF). Chr, Chromosome. G, Subcellular localization of NRL9 protein. D53-mCherry was used as a nuclear localization marker. Scale bar is 200 μm. GFP, Green fluorescent protein. H, Expression pattern of NRL9 in different tissues. Rice seeds were sampled at the heading stage (25 d after flowering). Gene expression was measured by qRT-PCR using the Ubiquitin gene as the internal control. I, Phenotypes of panicles, grain length, and grain width of WT and NRL9-overexpressing lines. Scale bars are 1 cm. J, Grain length, grain width, 1000-grain weight, and seed-setting rate for WT and NRL9-overexpressing lines. K, Phenotypes of grains, brown rice, and fracture surfaces of WT and NRL9-overexpressing lines. Scale bars are 1, 0.5, and 0.2 cm from the upper to lower panel. Values in B, E, H, and J represent Mean ± SD (n = 3). Statistically significant differences between mutants and WT were determined in B and E using the Student’s t-test (**, P < 0.01), and analyzed in J using the LSD method for multiple comparisons (different lowercase letters indicate statistically significant differences at P < 0.05).

Fig. 1. NARROW AND ROLLED LEAF 9 (NRL9) is a novel gene regulating leaf morphology and grain size in rice. A, Plant and leaf morphologies at the heading stage, and panicle and grain morphologies at the maturity stage of wild type (WT) and nrl9 mutant. Scale bars are 10, 1, 2, 1, and 1 cm from left to right, respectively. B, Leaf width, leaf rolling index, grain length, and grain width for WT and nrl9 mutants. C, Cytological analysis of leaf cross-sections for WT and nrl9 mutants. LV, Large vein; SV, Small vein; SC(ab), Sclerenchyma cells (abaxial). The yellow and red boxes indicate the normal development of sclerenchyma cells and the partially developed defective sclerenchyma cells, respectively. The numbers in the figure indicate the sequential count of SVs between two LVs. Scale bars are 1000, 100, and 50 μm from left to right, respectively. D, Morphology of non-flowered spikelet hulls. The red and blue boxes indicate the local enlarged views of the lemma and palea surfaces of the glume wax sections, respectively. Scale bars are 1000, 500, 50, and 50 μm from left to right, respectively. E, Number of large and small veins in WT and nrl9 mutant leaves, as well as cell lengths of lemma, palea, and total. F, Map-based cloning of NRL9 gene. NRL9 was mapped to a 110-kb interval on chromosome 9. Sequencing revealed a non-synonymous mutation from G to A in exon of the 6th open reading frame (ORF). Chr, Chromosome. G, Subcellular localization of NRL9 protein. D53-mCherry was used as a nuclear localization marker. Scale bar is 200 μm. GFP, Green fluorescent protein. H, Expression pattern of NRL9 in different tissues. Rice seeds were sampled at the heading stage (25 d after flowering). Gene expression was measured by qRT-PCR using the Ubiquitin gene as the internal control. I, Phenotypes of panicles, grain length, and grain width of WT and NRL9-overexpressing lines. Scale bars are 1 cm. J, Grain length, grain width, 1000-grain weight, and seed-setting rate for WT and NRL9-overexpressing lines. K, Phenotypes of grains, brown rice, and fracture surfaces of WT and NRL9-overexpressing lines. Scale bars are 1, 0.5, and 0.2 cm from the upper to lower panel. Values in B, E, H, and J represent Mean ± SD (n = 3). Statistically significant differences between mutants and WT were determined in B and E using the Student’s t-test (**, P < 0.01), and analyzed in J using the LSD method for multiple comparisons (different lowercase letters indicate statistically significant differences at P < 0.05).

 ), Hu Peisong(
), Hu Peisong( )
)