
Rice Science ›› 2021, Vol. 28 ›› Issue (4): 313-316.DOI: 10.1016/j.rsci.2021.05.001
• Letter • Next Articles
Yachun Yang1,2, Juan Li2, Hao Li2, Zuntao Xu3, Ruiying Qin2, Wenge Wu2, Pengcheng Wei2, Yong Ding3( ), Jianbo Yang2(
), Jianbo Yang2( )
)
Received:2020-07-31
Accepted:2020-11-18
Online:2021-07-28
Published:2021-07-28
Yachun Yang, Juan Li, Hao Li, Zuntao Xu, Ruiying Qin, Wenge Wu, Pengcheng Wei, Yong Ding, Jianbo Yang. SDF5 Encoding P450 Protein Is Required for Internode Elongation in Rice[J]. Rice Science, 2021, 28(4): 313-316.
Add to citation manager EndNote|Ris|BibTeX
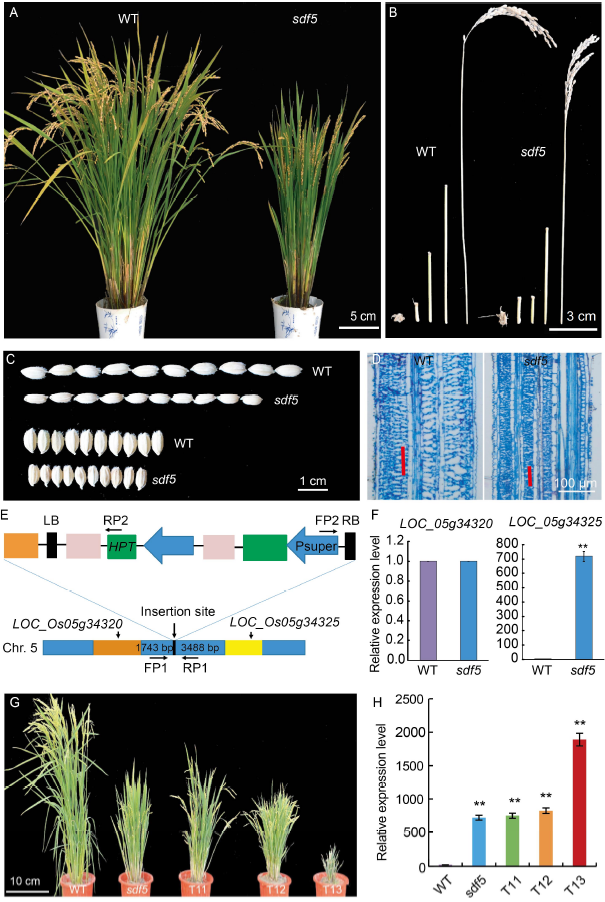
Fig. 1. Characterization of sdf5A, Phenotypes of the wild type (WT) and the sdf5 mutant. B, Internodes of WT and the sdf5 mutant. C, Grain size of WT and the sdf5 mutant. D, Cell length of the fifth internode in WT and the sdf5 mutant. E, Diagram of T-DNA insertion in the sdf5 mutant. FP1, FP2, RP1, and RP2 indicate the primers used for genotyping. F, Transcripts of LOC_ Os05g34320 and LOC_Os05g34325. G, Phenotypes of wild type, sdf5 mutant and LOC_Os05g34325 overexpression lines. T11, T12 and T13 indicate different overexpression plants. H, Transcripts of SDF5 in wild type, sdf5, T11, T12 and T13. Data are Mean ± SD (n = 3). * and ** indicate significant differences at the 0.05 and 0.01 levels by the Student’s t-test, respectively.
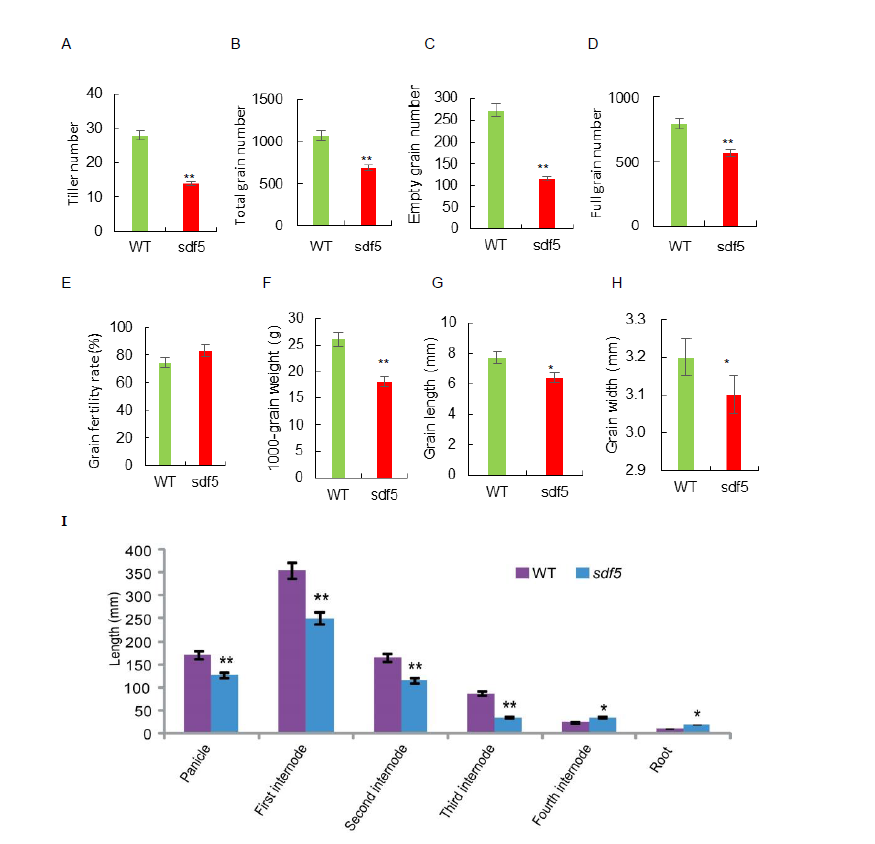
Fig. S1. Agronomic traits of wild type (WT) and sdf5 mutant. A, Tiller number. B, Total grain number per panicle. C, Empty grain number per panicle. D, Full grain number per panicle. E, Grain fertility rate. F, 1000-grain weight; G, Grain length. H, Grain width. I, Internode length. All agronomic character surveys were conducted after the rice harvest. Data are Mean ± SD (n = 3). * and ** indicate significant differences at the 0.05 and 0.01 levels by the Student’s t-test, resppectively.
| Generation | Total number of plants | Number of normal plants | Number of semi-dwarf | χ2 |
|---|---|---|---|---|
| T1 | 21 | 7 | 14 | 0.38 |
| T2 | 420 | 178 | 242 | 0.07 |
Table S1. Genetic analysis of SDF5.
| Generation | Total number of plants | Number of normal plants | Number of semi-dwarf | χ2 |
|---|---|---|---|---|
| T1 | 21 | 7 | 14 | 0.38 |
| T2 | 420 | 178 | 242 | 0.07 |
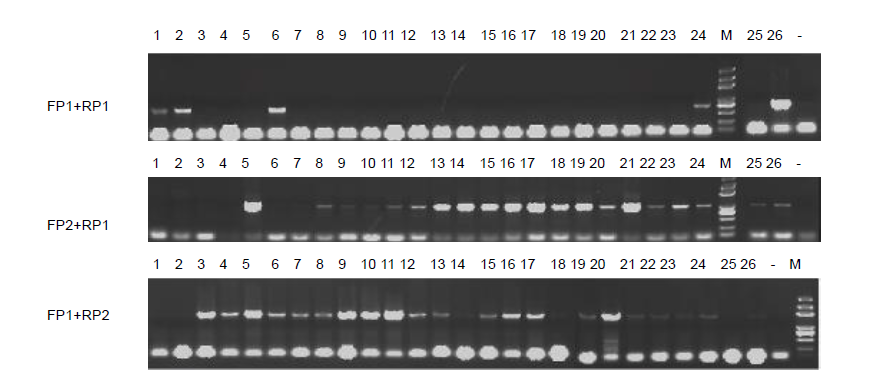
Fig. S2. Segregation of SDF5 in F2 population with self-pollilation. FP1 and RP1 indicate the left primer and right primer on T-DNA border; FP2 is a primer of the Psuper primer; RP2 is a primer in the HPT sequence. Lanes 1 and 2 are wild type. Lanes 3 to 26 are SDF5. ‘-’ indicates the blank. M, Marker.
| Primer name | Primer sequence (5'-3') | Primer name | Primer sequence (5'-3') |
|---|---|---|---|
| FP1 | CAATAAGGAGCAGGGCCCAGT | SDF5-qRT-F | ATTGCATCGCTCTCCTAGGGC |
| FP2 | AGGACGACCAATGTATGTTATG | SDF5-qRT-R | GCCGGATTGCTGAAGGCGACG |
| RP1 | ACAAGTATGGAAACTACTAGA | LRT2-qRT-F1 | CTACAAGGGGAGCACCTTCCA |
| RP2 | CATGTCATATAGTTTCCATAGC | LRT2-qRT-F2 | TGTACGCGAAGGACGTGCCG |
| 34325F CDS 1300 KpnI | GGTACCATGGAGTTAACTAGTAGCAGTGCCA | LRT2-qRT-R1/2 | GAAGAACTGGGACCCGTTAGT |
| 34325R CDS 1300 XbaI | CTCTAGAAGAGGTAGGAAGCAATTGCCTTCT | OsActin2-qRT-F | TACAGTGTCTGGATAGGAGGGTC |
| 34320F CDS 1300 BamHI | GGATCCATGGCGCCAGCGATGGCTCTGAGGT | OsActin2-qRT-R | ACCAACAATCCCAAACAGAGTAG |
| 34320R CDS 1300 XbaI | CTCTAGCTGCTTTATACAGGAGCTTGGCTCT | OsIAA1-qRT-F | CGAGGCCTACTCCGGCTACGAC |
| SDF5F CDS GFP XbaI | CTCTAGATGGAGTTAACTAGTAGCAGTGCCA | OsIAA1-qRT-R | CGCTGGCAAGTTTCCACAAACA |
| SDF5R CDS GFP KpnI | GGTACCAAGAGGTAGGAAGCAATTGCCTTCT | OsIAA5-qRT-F | GAACTACCGAAAGAACACGCTG |
| OsKO2-qRT-F | TGCTACCAGCGACTATTGTGATTT | OsIAA5-qRT-R | TTTGATGATTTACGCAAACCGC |
| OsKO2-qRT-R | GTGCAGAAGTACCCAACATGCTT | OsIAA10-qRT-F | ATGAGAGGAGGAGTAGCTGGGC |
| OsKO-qRT-F | CTTCCTCCATCATTTTCTCC | OsIAA10-qRT-R | TCAGGATCTGCCTCTTGTTG |
| OsKO-qRT-R | AAGCAGTTGTCCACAGGC | OsIAA11-qRT-F1 | TGTTCACCTCCTTCACCAT |
| OsGA20ox2-qRT-F | CCAATTTTGGACCCTACCGC | OsIAA11-qRT-F2 | CCGAGACCATCGACCTCAA |
| OsGA20ox2-qRT-R | GAGAGAAGCCCAACCCAACC | OsIAA11-qRT-R1 | GCATTTCTCCACTGCCCTT |
| OsGA2ox1-qRT-F | CGTCTTGTAGATGGTGGTGC | OsIAA11-qRT-R2 | CGGCCACCCCACCACCTGC |
| OsGA2ox1-qRT-R | CCTGCCTGATGAGTTAGAAAAG | OsIAA12-qRT-F | GCAACAACAAAGACGAGCCCAT |
| OsGA2ox3-qRT-F | TGGTGGCCAACAGCCTAAAG | OsIAA12-qRT-R | CCTTGTCCTGGTAGGTGATGGC |
| OsGA2ox3-qRT-R | TGGTGCAATCCTCTGTGCTAAC | OsIAA13-qRT-F | AGCATGGACGGGGCACCCTACCT |
| GID1-qRT-F | GCCCATCCTTGAGTTCCTGAC | OsIAA13-qRT-R | TCTGCCACGGGACATCTCCAACA |
| GID1-qRT-F | GTCCTGGAGCGTCACGAAGTA | OsIAA14-qRT-F | GAGGAGGGCTGTACGTGAAGGTG |
| GA3OX2-qRT-F | CGGACTCGGGCTTCTTCACC | OsIAA14-qRT-R | AGAGATGAACATGTCCCAGGGCA |
| GA3OX2-qRT-R | AGGAAGTAGCCGAGCGAGACCC | OsIAA16-qRT-F | GACGACGACGACGAGGCGAGGT |
| GA2OX1-qRT-F | TACCTGCTCCTCCACGCCAACC | OsIAA16-qRT-R | GCCGCCGGAGAAGCAGAGGAACAT |
| GA2OX1-qRT-R | CACTGTCAGCGTTTGTGGTAAG | OsIAA20-qRT-F | CGGTGGGTTCAAAACCGACGA |
| KO1-qRT-F | AAGCAGCAGAAGGAAAGG | OsIAA20-qRT-R | GCTCAGCGTATGAGCCGAGGA |
| KO1-qRT-R | TGGTAAAGCCGTTCCTGT | OsIAA22-qRT-F | TCATCAGCTCAGCTGGTGGGATG |
| OsGID2-qRT-F | GGGAAATGACAGTGATAAGAATGGA | OsIAA22-qRT-R | AGAGCCTGGGACAGCGAGTCGTA |
| OsGID2-qRT-R | TGATGCTTGTTGGAGTGAAGCT | OsIAA30-qRT-F | AAGATGTACAAGAGCTACCTTGA |
| OsSLR1-qRT-F | GACGTCAACGAACGCTCAATT | OsIAA30-qRT-R | TTCTGATCCTTTCATTATCCTAA |
| OsSLR1-qRT-R | CGGAGTCCAGTCGTCGATCT | OsIAA31-qRT-F | CGGAGACGCAGCAGAAGGAGGATG |
| OsActin-F | GCCTTGGCAATCCACATC | OsIAA31-qRT-R | GTAGGTGACGGCGAAGTCGGACGG |
| OsActin-R | AGCATGAAGATCAAGGTGGTC |
Table S2. Primers used for gene over-expression, PCR and RT-PCR analysis.
| Primer name | Primer sequence (5'-3') | Primer name | Primer sequence (5'-3') |
|---|---|---|---|
| FP1 | CAATAAGGAGCAGGGCCCAGT | SDF5-qRT-F | ATTGCATCGCTCTCCTAGGGC |
| FP2 | AGGACGACCAATGTATGTTATG | SDF5-qRT-R | GCCGGATTGCTGAAGGCGACG |
| RP1 | ACAAGTATGGAAACTACTAGA | LRT2-qRT-F1 | CTACAAGGGGAGCACCTTCCA |
| RP2 | CATGTCATATAGTTTCCATAGC | LRT2-qRT-F2 | TGTACGCGAAGGACGTGCCG |
| 34325F CDS 1300 KpnI | GGTACCATGGAGTTAACTAGTAGCAGTGCCA | LRT2-qRT-R1/2 | GAAGAACTGGGACCCGTTAGT |
| 34325R CDS 1300 XbaI | CTCTAGAAGAGGTAGGAAGCAATTGCCTTCT | OsActin2-qRT-F | TACAGTGTCTGGATAGGAGGGTC |
| 34320F CDS 1300 BamHI | GGATCCATGGCGCCAGCGATGGCTCTGAGGT | OsActin2-qRT-R | ACCAACAATCCCAAACAGAGTAG |
| 34320R CDS 1300 XbaI | CTCTAGCTGCTTTATACAGGAGCTTGGCTCT | OsIAA1-qRT-F | CGAGGCCTACTCCGGCTACGAC |
| SDF5F CDS GFP XbaI | CTCTAGATGGAGTTAACTAGTAGCAGTGCCA | OsIAA1-qRT-R | CGCTGGCAAGTTTCCACAAACA |
| SDF5R CDS GFP KpnI | GGTACCAAGAGGTAGGAAGCAATTGCCTTCT | OsIAA5-qRT-F | GAACTACCGAAAGAACACGCTG |
| OsKO2-qRT-F | TGCTACCAGCGACTATTGTGATTT | OsIAA5-qRT-R | TTTGATGATTTACGCAAACCGC |
| OsKO2-qRT-R | GTGCAGAAGTACCCAACATGCTT | OsIAA10-qRT-F | ATGAGAGGAGGAGTAGCTGGGC |
| OsKO-qRT-F | CTTCCTCCATCATTTTCTCC | OsIAA10-qRT-R | TCAGGATCTGCCTCTTGTTG |
| OsKO-qRT-R | AAGCAGTTGTCCACAGGC | OsIAA11-qRT-F1 | TGTTCACCTCCTTCACCAT |
| OsGA20ox2-qRT-F | CCAATTTTGGACCCTACCGC | OsIAA11-qRT-F2 | CCGAGACCATCGACCTCAA |
| OsGA20ox2-qRT-R | GAGAGAAGCCCAACCCAACC | OsIAA11-qRT-R1 | GCATTTCTCCACTGCCCTT |
| OsGA2ox1-qRT-F | CGTCTTGTAGATGGTGGTGC | OsIAA11-qRT-R2 | CGGCCACCCCACCACCTGC |
| OsGA2ox1-qRT-R | CCTGCCTGATGAGTTAGAAAAG | OsIAA12-qRT-F | GCAACAACAAAGACGAGCCCAT |
| OsGA2ox3-qRT-F | TGGTGGCCAACAGCCTAAAG | OsIAA12-qRT-R | CCTTGTCCTGGTAGGTGATGGC |
| OsGA2ox3-qRT-R | TGGTGCAATCCTCTGTGCTAAC | OsIAA13-qRT-F | AGCATGGACGGGGCACCCTACCT |
| GID1-qRT-F | GCCCATCCTTGAGTTCCTGAC | OsIAA13-qRT-R | TCTGCCACGGGACATCTCCAACA |
| GID1-qRT-F | GTCCTGGAGCGTCACGAAGTA | OsIAA14-qRT-F | GAGGAGGGCTGTACGTGAAGGTG |
| GA3OX2-qRT-F | CGGACTCGGGCTTCTTCACC | OsIAA14-qRT-R | AGAGATGAACATGTCCCAGGGCA |
| GA3OX2-qRT-R | AGGAAGTAGCCGAGCGAGACCC | OsIAA16-qRT-F | GACGACGACGACGAGGCGAGGT |
| GA2OX1-qRT-F | TACCTGCTCCTCCACGCCAACC | OsIAA16-qRT-R | GCCGCCGGAGAAGCAGAGGAACAT |
| GA2OX1-qRT-R | CACTGTCAGCGTTTGTGGTAAG | OsIAA20-qRT-F | CGGTGGGTTCAAAACCGACGA |
| KO1-qRT-F | AAGCAGCAGAAGGAAAGG | OsIAA20-qRT-R | GCTCAGCGTATGAGCCGAGGA |
| KO1-qRT-R | TGGTAAAGCCGTTCCTGT | OsIAA22-qRT-F | TCATCAGCTCAGCTGGTGGGATG |
| OsGID2-qRT-F | GGGAAATGACAGTGATAAGAATGGA | OsIAA22-qRT-R | AGAGCCTGGGACAGCGAGTCGTA |
| OsGID2-qRT-R | TGATGCTTGTTGGAGTGAAGCT | OsIAA30-qRT-F | AAGATGTACAAGAGCTACCTTGA |
| OsSLR1-qRT-F | GACGTCAACGAACGCTCAATT | OsIAA30-qRT-R | TTCTGATCCTTTCATTATCCTAA |
| OsSLR1-qRT-R | CGGAGTCCAGTCGTCGATCT | OsIAA31-qRT-F | CGGAGACGCAGCAGAAGGAGGATG |
| OsActin-F | GCCTTGGCAATCCACATC | OsIAA31-qRT-R | GTAGGTGACGGCGAAGTCGGACGG |
| OsActin-R | AGCATGAAGATCAAGGTGGTC |
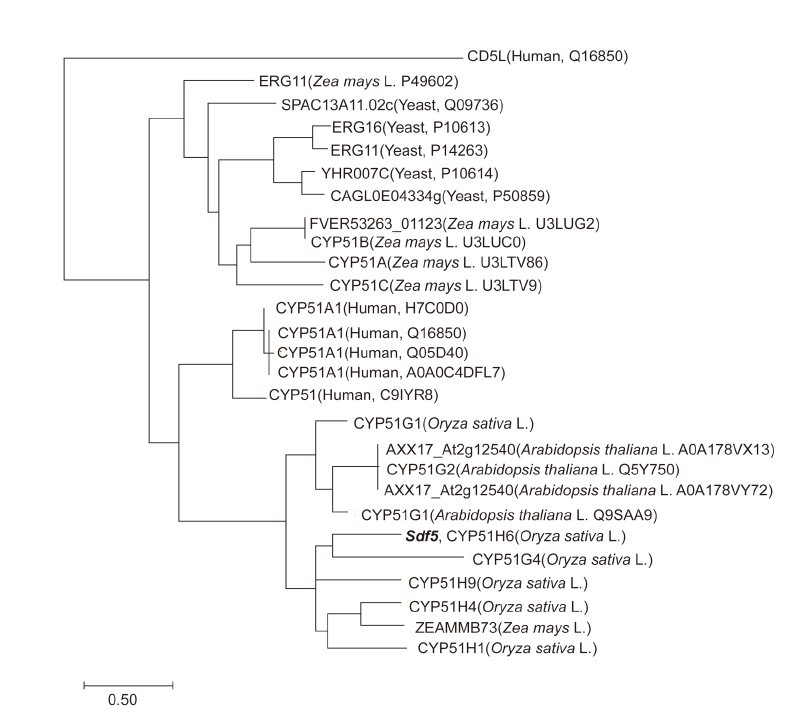
Fig. S3. Phylogenetic tree of CYP51 proteins. Cluster analysis using the protein sequences of the CYP51 proteins of rice, yeast, human and Arabidopsis thaliana aligned by ClustalW. The relationships of the sequences were examined by MEGA-X.

Fig. S4. Expression pattern of SDF5. Total RNA was isolated from 8-week-old seedlings. Data are Mean ± SD (n = 3).** indicate significant difference by Student’s t-test. P < 0.01.
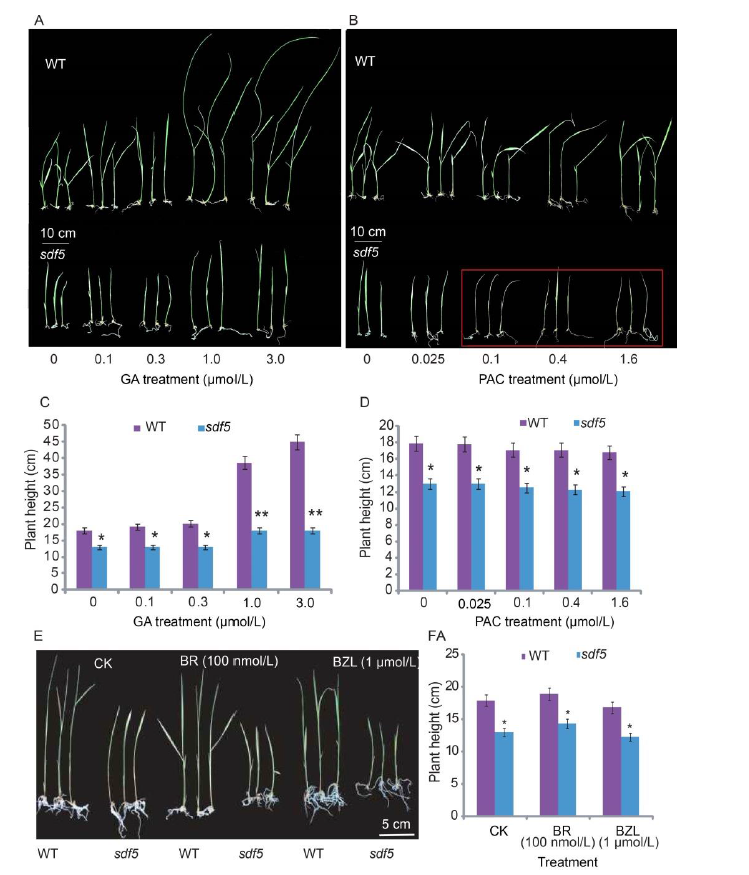
Fig. S5. Wild type (WT) and sdf5 mutants treated with gibberellic acid (GA3), paclobutrazol (PAC) and brassinosteriod (BR). A and C, Plant height with GA3 treatment. B and D, Plant height with PAC treatment. Red frame marks seedling death. E, Phonetype of wild type and sdf5 treated with BR and benzoyl-CoA ligase (BZL). F, Plant height of WT and sdf5 mutant with BR and BZL treatments. 15-day-old seedlings were subjected to 10 d. Data are Mean ± SD (n = 3). * and ** indicate significant differences at the 0.05 and 0.01 levels by the Student’s t-test, resppectively. CK, Control.
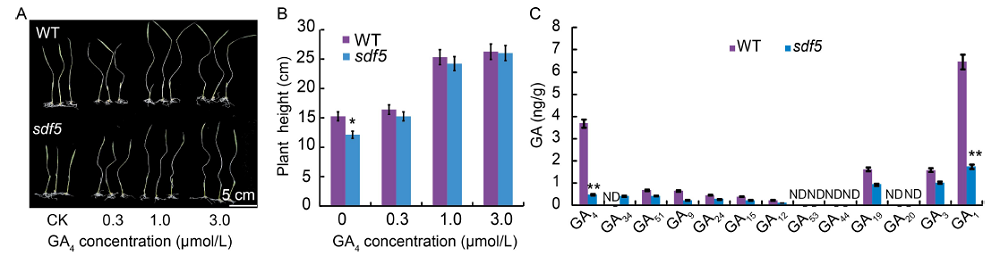
Fig. 2. Gibberellin (GA) compounds were tested in wild type (WT) and sdf5 mutant. A, Phenotypes of WT and sdf5 mutant treated with different concentrations of GA4. Fifteen-day-old seedlings were subjected to 10 d of homone culture and their plant heights were measured. CK, Control. B, Height of WT and sdf5 seedlings in different concentrations of GA4. C, Endogenous levels of GAs were measured in WT and sdf5 mutant. ND indicates not detectable. Data are Mean ± SD (n = 3). * and ** indicate significant differences at the 0.05 and 0.01 levels by the Student’s t-test, respectively.

Fig. S6. Transcriptional analysis of GA genes in wild type (WT) and sdf5 mutant. Total RNA was isolated from 2-week-old seedlings. Data are Mean ± SD (n = 3). * indicate significant differences at the 0.05 level by the Student’s t-test.
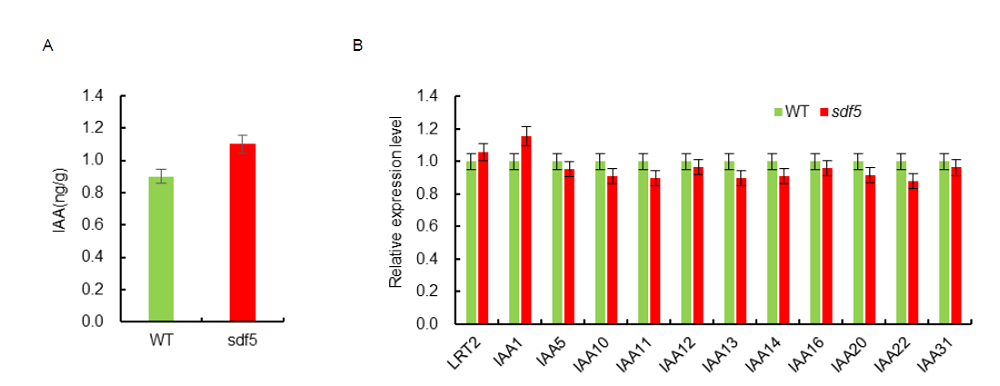
Fig. S7. Indoleacetic acid (IAA) analysis in wild type (WT) and sdf5 mutant. A, IAA content in WT and sdf5 mutant. IAA content was tested with 2-week-old seedlings. B, Transcript level of IAA genes in sdf5 mutant. Total RNA was isolated from 2-week-old seedlings. Data are Mean ± SD (n = 3).
| [1] | Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. 2009. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol, 50(8): 1416-1424. |
| [2] | Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim R B, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M. 2009. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol Genet Genom, 281: 223-231. |
| [3] | Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. 1999. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci USA, 96(18): 10284-10289. |
| [4] | Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y. 1999. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA, 96: 7575-7580. |
| [5] | Jiang L, Liu X, Xiong G S, Liu H H, Chen F L, Wang L, Meng X B, Liu G F, Yu H, Yuan Y D, Yi W, Zhao L H, Ma H L, He Y Z, Wu Z S, Melcher K, Qian Q, Xu H E, Wang Y H, Li J Y. 2013. DWARF53 acts as a repressor of strigolactone signalling in rice. Nature, 504: 401-405. |
| [6] | Liang F, Xin X Y, Hu Z J, Xu J D, Wei G, Qian X Y, Yang J S, He H H, Luo X J. 2011. Genetic analysis and fine mapping of a novel semidominant dwarfing gene LB4D in rice. J Integr Plant Biol, 53(4): 312-323. |
| [7] | Lin H, Wang R X, Qian Q, Yan M X, Meng X B, Fu Z M, Yan C Y, Jiang B, Su Z, Li J Y, Wang Y H. 2009. DWARF27, an iron- containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell, 21(5): 1512-1525. |
| [8] | Liu B M, Wu Y J, Fu X D, Qian Q. 2009. Characterizations and molecular mapping of a novel dominant semi-dwarf gene Sdd(t) in rice (Oryza sativa). Plant Breeding, 127(2): 125-130. |
| [9] | Magome H, Nomura T, Hanada A, Takeda-Kamiya N, Ohnishi T, Shinma Y, Katsumata T, Kawaide H, Kamiya Y, Yamaguchi S. 2013. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci USA, 110(5): 1947‒1952. |
| [10] | Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y. 2002. Positional cloning of rice semidwarfing gene, sd-1: Rice ‘green revolution gene’ encodes a mutant enzyme involved in gibberellin synthesis. DNA Res, 9(1): 11-17. |
| [11] | Osbourn A, Goss R J M, Field R A. 2011. The saponins: Polar isoprenoids with important and diverse biological activities. Nat Prod Rep, 28: 1261-1268. |
| [12] | Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush G S, Kitano H, Matsuoka M. 2002. A mutant gibberellin-synthesis gene in rice. Nature, 416(2): 701-702. |
| [13] | Spielmeyer W, Ellis M H, Chandler P M. 2002. Semidwarf (sd-1), ‘green revolution’ rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA, 99(13): 9043-9048. |
| [14] | Sun L J, Li X J, Fu Y C, Zhu Z F, Tan L B, Liu F X, Sun X Y, Sun X W, Sun C Q. 2013. GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J Integr Plant Biol, 55(5): 938-949. |
| [15] | Sunohara H, Kawai T, Shimizu-Sato S, Sato Y, Sato K, Kitano H. 2009. A dominant mutation of TWISTED DWARF 1 encoding an α-tubulin protein causes severe dwarfism and right helical growth in rice. Genes Genet Syst, 84(3): 209-218. |
| [16] | Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M. 2000. Rice dwarf mutant d1, which is defective in the α subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA, 97(21): 11638-11643. |
| [17] | Wang X, Yu H X, Tang D, Huang J, Gong Z Y, Cheng Z K. 2008. Genetic analysis of a dominant dwarf mutant in rice (Oryza sativa L.). Sci Agric China, 41: 3959‒3966. (in Chinese with English abstract) |
| [18] | Xia K F, Ou X J, Tang H D, Wang R, Wu P, Jia Y X, Wei X Y, Xu X L, Kang S H, Kim S K, Zhang M Y. 2015. Rice microRNA osa-miR1848 targets the obtusifoliol 14α-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytol, 208(3): 790-802. |
| [19] | Yan H F, Saika H, Maekawa M, Takamure I, Tsutsumi N, Kyozuka J, Nakazono M. 2007. Rice tillering dwarf mutant dwarf3 has increased leaf longevity during darkness-induced senescence or hydrogen peroxide-induced cell death. Genes Genet Syst, 82(4): 361-366. |
| [20] | Zhang S Y, Li G, Fang J, Chen W Q, Jiang H P, Zou J H, Liu X, Zhao X F, Li X B, Chu C C, Xie Q, Jiang X N, Zhu L H. 2010. The interactions among DWARF10, auxin and cytokinin underlie lateral bud outgrowth in rice. J Integr Plant Biol, 52(7): 626-638. |
| [21] | Zhang L G, Cheng Z J, Qin R Z, Qiu Y, Wang J L, Cui X K, Gu L F, Zhang X, Guo X P, Wang D, Jiang L, Wu C Y, Wang H Y, Cao X F, Wan J M. 2012. Identification and characterization of an Epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell, 24(11): 4407-4421. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||