
Rice Science ›› 2021, Vol. 28 ›› Issue (4): 322-324.DOI: 10.1016/j.rsci.2021.05.003
• Letter • Previous Articles Next Articles
Riaz Aamir1,#, Huimin Wang1,#, Zhenhua Zhang1, Anley Zegeye Workie1,2, Yanhui Li1, Hong Wang1, Pao Xue1, Zequn Peng1, Xihong Shen1, Shihua Cheng1( ), Yingxin Zhang1(
), Yingxin Zhang1( )
)
Received:2020-07-20
Accepted:2020-11-13
Online:2021-07-28
Published:2021-07-28
About author:#These authors contributed equally to this work
Riaz Aamir, Huimin Wang, Zhenhua Zhang, Anley Zegeye Workie, Yanhui Li, Hong Wang, Pao Xue, Zequn Peng, Xihong Shen, Shihua Cheng, Yingxin Zhang. Development of Chromosome Segment Substitution Lines and Genetic Dissection of Grain Size Related Locus in Rice[J]. Rice Science, 2021, 28(4): 322-324.
Add to citation manager EndNote|Ris|BibTeX
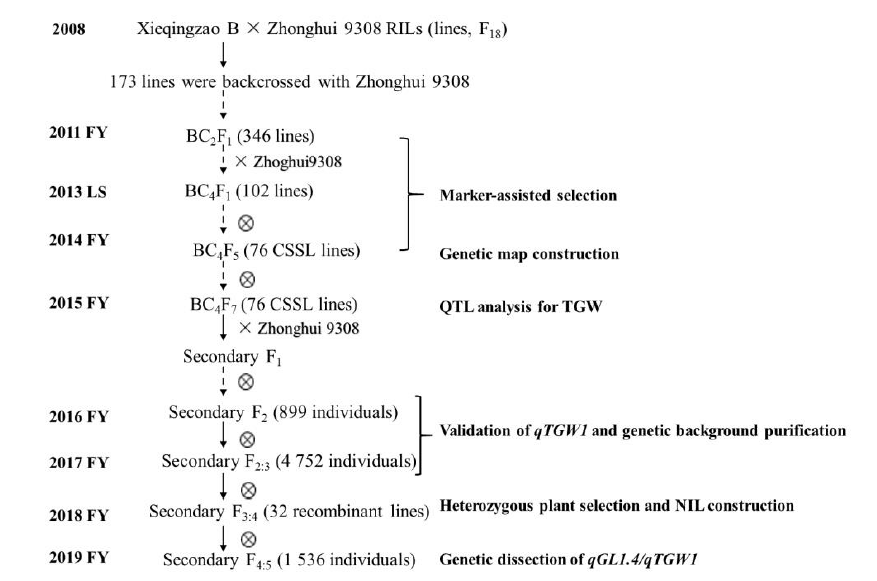
Fig. S1. Detailed scheme for population development. FY, Fuyang, Zhejiang Province, China; LS, Lingshui, Hainan Province, China; CSSL, Chromosome segment substitution line; RIL, Recombinant inbred line; TGW, 1000-grain weight.
| Name | Chr. | Forward | Reverse | Purpose |
|---|---|---|---|---|
| InDel1 | 1 | AGGGGAAGAAAAACCTGACC | CCGCGTGCAGATAAAGTACA | Linkage map |
| InDel7 | 1 | TGGCCCAATAGCCCATTTAT | CGAGAGCCCGAGAGAGAGA | Linkage map |
| InDel15 | 1 | CAACCCCTCCAAATACCTGA | ACCGTGTTCATGCCTTTCAC | Linkage map |
| InDel21 | 2 | ATAGGGTGGGTGTGCTGAAC | GCACAAAACTGCAGGTCTCC | Linkage map |
| InDel24 | 2 | TACCTCGGCTCGGGTCAAT | CGACCAAGCGAGAAGGTACT | Linkage map |
| InDel28 | 2 | GGCTGGCTGTTGCTCATC | AAAAATCCCAACCCTGCTG | Linkage map |
| InDel31 | 2 | GGAAGCTTCAGCCTCACG | GGTTATACAACGGCGGATCT | Linkage map |
| InDel36 | 3 | TGGTTTATATTGGAACGGAGGA | GTTACATGCCCTTTCGCAGT | Linkage map |
| InDel51 | 3 | CCATCTCTTTCCACGACGAT | AGTGCGGCGAACAGATAAAG | Linkage map |
| InDel55 | 3 | AGGTCTCGTGTCGTTCATCC | TGGAGGGAGCATGTCTATCA | Linkage map |
| InDel62 | 4 | CCACGTATAAAGCAGTTTGTTAGG | CAAGTGTGGTTGTTAGCATCAAA | Linkage map |
| InDel65 | 4 | GCGTACAGCGAGAGGTTGAC | TCTCTTCGCCACGGAGAC | Linkage map |
| InDel71 | 4 | ATGTAACCCGGCCAGAGTG | CCATTAACTGGTCGGAATCG | Linkage map |
| InDel78 | 5 | AAATTTAGGCCAGGCAGCTT | TCTCTCACACGCTTATTCATCTTT | Linkage map |
| InDel79 | 5 | CGTGCCGATGACAAACTTC | GAGGATCCATGTCCACCATT | Linkage map |
| InDel84 | 5 | TGAGTTTCCGGTGTTCCATA | AAGGCAAAGTCGTTCAGCTT | Linkage map |
| InDel85 | 6 | GCAATCTAGTAAACTGTTCGAGAAA | TGGAATTTAAACATCCTCAATGC | Linkage map |
| InDel90 | 6 | CCTCATCCAGGGGTCATGTA | CGGTCAAGTGTCATCCAGGT | Linkage map |
| InDel94 | 6 | GGCATTGTAGCCAATCCAGA | AAACACACTCCCCCATGAGA | Linkage map |
| InDel98 | 7 | TGACTGTTACCCTTACGTGCAG | CGGGATGAAACAGATTCTGAG | Linkage map |
| InDel103 | 7 | CCCCATGAGGCCTACACTT | AGCAGCATAATCAGATGAGACG | Linkage map |
| InDel108 | 7 | GCCCACCTGTCATTGAGAGTA | GTTTTTGCGCTTTTGTTGCT | Linkage map |
| InDel113 | 8 | TTTTAAAGCTGCGCCAAAAG | CATAACCGGTAAAGGAGTAGCC | Linkage map |
| InDel121 | 8 | AACCATGAATGAATCCCTGA | TGCAACTGACATCCTGCAAT | Linkage map |
| InDel126 | 9 | GCCGGCCTTATCCATTTTT | GAGCGCCACTGCTTCTACTC | Linkage map |
| InDel129 | 9 | GCGAACCGATAAAACTGCTC | AGAGGTGTATCAAAGCAATCGAG | Linkage map |
| InDel133 | 10 | AATTCTTATGGACGGATACGC | TCAGCATCTCGTAAGCAAAAA | Linkage map |
| InDel135 | 10 | TTTCTCCTTTCATCCACTGCT | AACGTGGAACCCTAGTCAAGAA | Linkage map |
| InDel144 | 11 | TGATGAGCTCTCACTTGTTGAAA | CGTACATTGGCTTATGTGATCTG | Linkage map |
| InDel151 | 11 | TGCAGTACAACACTCAGTTCAAA | CATGTTACGGTACTGGCATCA | Linkage map |
| InDel156 | 12 | TCTCAAGCATGTCAAGGCTTA | ATGAACATGCAGAGCACCAA | Linkage map |
| InDel159 | 12 | TGGGCAACTGAATCTAACCA | GGAGATGATGATGCGGTGAT | Linkage map |
| InDel165 | 12 | TCAGACACAACGTACACATCG | TCGATTGATCACTGACGGTTA | Linkage map |
| IN3-3 | 3 | GCATGCGTAGCTAGTAAAACTT | AACGTACTGTTGCTGCCTTA | Genetic dissection |
| IN3-9 | 3 | CACCCACCATGATCCAAGAA | CGAAACGCTCGTTATATCTCC | Genetic dissection |
| IN3-14 | 3 | CAGGTAGTGGTGGGCTTCTT | ACCAAAATCCCTCGAGAACT | Genetic dissection |
| IN3-22 | 3 | TTTGTTGGTTGTCTCCTCCG | GTCGTGGTCCTCGTCCAT | Genetic dissection |
| IN3-25 | 3 | TGCAAGTAAATACGAACCTCA | CATTTTATGATGCGTGGGCA | Genetic dissection |
| IN7-1 | 7 | TATTCTTTTCTGTATCCGGCCC | GCGTGGAAGAAAACAACGG | Genetic dissection |
| IN7-7 | 7 | CACATCCATCGCACTCTTGA | GCCTATTGCCCATTACGTCT | Genetic dissection |
| IN7-12 | 7 | CAGTGAGAGAAGAGAGCCAAG | AGCCGTCGTCGATCAAGTT | Genetic dissection |
| IN7-14 | 7 | TCAAATTCCAGGTTGCACTGT | AGTGTGTCATGTTGCAGTCATA | Genetic dissection |
| IN7-18 | 7 | AAGCTTTCGGTCAACAACAT | ATGCCCCATATCCAGATTCAG | Genetic dissection |
| D-12 | 1 | AAAAGCATCGAATCGCGAAA | TTGATTCATTGTTTGGGGCA | Fine mapping |
| TG-57 | 1 | CCACTAGTTGGAGCAGACAC | CGTCTTCCGGAAATTGTTGG | Fine mapping |
Table S1. List of primers.
| Name | Chr. | Forward | Reverse | Purpose |
|---|---|---|---|---|
| InDel1 | 1 | AGGGGAAGAAAAACCTGACC | CCGCGTGCAGATAAAGTACA | Linkage map |
| InDel7 | 1 | TGGCCCAATAGCCCATTTAT | CGAGAGCCCGAGAGAGAGA | Linkage map |
| InDel15 | 1 | CAACCCCTCCAAATACCTGA | ACCGTGTTCATGCCTTTCAC | Linkage map |
| InDel21 | 2 | ATAGGGTGGGTGTGCTGAAC | GCACAAAACTGCAGGTCTCC | Linkage map |
| InDel24 | 2 | TACCTCGGCTCGGGTCAAT | CGACCAAGCGAGAAGGTACT | Linkage map |
| InDel28 | 2 | GGCTGGCTGTTGCTCATC | AAAAATCCCAACCCTGCTG | Linkage map |
| InDel31 | 2 | GGAAGCTTCAGCCTCACG | GGTTATACAACGGCGGATCT | Linkage map |
| InDel36 | 3 | TGGTTTATATTGGAACGGAGGA | GTTACATGCCCTTTCGCAGT | Linkage map |
| InDel51 | 3 | CCATCTCTTTCCACGACGAT | AGTGCGGCGAACAGATAAAG | Linkage map |
| InDel55 | 3 | AGGTCTCGTGTCGTTCATCC | TGGAGGGAGCATGTCTATCA | Linkage map |
| InDel62 | 4 | CCACGTATAAAGCAGTTTGTTAGG | CAAGTGTGGTTGTTAGCATCAAA | Linkage map |
| InDel65 | 4 | GCGTACAGCGAGAGGTTGAC | TCTCTTCGCCACGGAGAC | Linkage map |
| InDel71 | 4 | ATGTAACCCGGCCAGAGTG | CCATTAACTGGTCGGAATCG | Linkage map |
| InDel78 | 5 | AAATTTAGGCCAGGCAGCTT | TCTCTCACACGCTTATTCATCTTT | Linkage map |
| InDel79 | 5 | CGTGCCGATGACAAACTTC | GAGGATCCATGTCCACCATT | Linkage map |
| InDel84 | 5 | TGAGTTTCCGGTGTTCCATA | AAGGCAAAGTCGTTCAGCTT | Linkage map |
| InDel85 | 6 | GCAATCTAGTAAACTGTTCGAGAAA | TGGAATTTAAACATCCTCAATGC | Linkage map |
| InDel90 | 6 | CCTCATCCAGGGGTCATGTA | CGGTCAAGTGTCATCCAGGT | Linkage map |
| InDel94 | 6 | GGCATTGTAGCCAATCCAGA | AAACACACTCCCCCATGAGA | Linkage map |
| InDel98 | 7 | TGACTGTTACCCTTACGTGCAG | CGGGATGAAACAGATTCTGAG | Linkage map |
| InDel103 | 7 | CCCCATGAGGCCTACACTT | AGCAGCATAATCAGATGAGACG | Linkage map |
| InDel108 | 7 | GCCCACCTGTCATTGAGAGTA | GTTTTTGCGCTTTTGTTGCT | Linkage map |
| InDel113 | 8 | TTTTAAAGCTGCGCCAAAAG | CATAACCGGTAAAGGAGTAGCC | Linkage map |
| InDel121 | 8 | AACCATGAATGAATCCCTGA | TGCAACTGACATCCTGCAAT | Linkage map |
| InDel126 | 9 | GCCGGCCTTATCCATTTTT | GAGCGCCACTGCTTCTACTC | Linkage map |
| InDel129 | 9 | GCGAACCGATAAAACTGCTC | AGAGGTGTATCAAAGCAATCGAG | Linkage map |
| InDel133 | 10 | AATTCTTATGGACGGATACGC | TCAGCATCTCGTAAGCAAAAA | Linkage map |
| InDel135 | 10 | TTTCTCCTTTCATCCACTGCT | AACGTGGAACCCTAGTCAAGAA | Linkage map |
| InDel144 | 11 | TGATGAGCTCTCACTTGTTGAAA | CGTACATTGGCTTATGTGATCTG | Linkage map |
| InDel151 | 11 | TGCAGTACAACACTCAGTTCAAA | CATGTTACGGTACTGGCATCA | Linkage map |
| InDel156 | 12 | TCTCAAGCATGTCAAGGCTTA | ATGAACATGCAGAGCACCAA | Linkage map |
| InDel159 | 12 | TGGGCAACTGAATCTAACCA | GGAGATGATGATGCGGTGAT | Linkage map |
| InDel165 | 12 | TCAGACACAACGTACACATCG | TCGATTGATCACTGACGGTTA | Linkage map |
| IN3-3 | 3 | GCATGCGTAGCTAGTAAAACTT | AACGTACTGTTGCTGCCTTA | Genetic dissection |
| IN3-9 | 3 | CACCCACCATGATCCAAGAA | CGAAACGCTCGTTATATCTCC | Genetic dissection |
| IN3-14 | 3 | CAGGTAGTGGTGGGCTTCTT | ACCAAAATCCCTCGAGAACT | Genetic dissection |
| IN3-22 | 3 | TTTGTTGGTTGTCTCCTCCG | GTCGTGGTCCTCGTCCAT | Genetic dissection |
| IN3-25 | 3 | TGCAAGTAAATACGAACCTCA | CATTTTATGATGCGTGGGCA | Genetic dissection |
| IN7-1 | 7 | TATTCTTTTCTGTATCCGGCCC | GCGTGGAAGAAAACAACGG | Genetic dissection |
| IN7-7 | 7 | CACATCCATCGCACTCTTGA | GCCTATTGCCCATTACGTCT | Genetic dissection |
| IN7-12 | 7 | CAGTGAGAGAAGAGAGCCAAG | AGCCGTCGTCGATCAAGTT | Genetic dissection |
| IN7-14 | 7 | TCAAATTCCAGGTTGCACTGT | AGTGTGTCATGTTGCAGTCATA | Genetic dissection |
| IN7-18 | 7 | AAGCTTTCGGTCAACAACAT | ATGCCCCATATCCAGATTCAG | Genetic dissection |
| D-12 | 1 | AAAAGCATCGAATCGCGAAA | TTGATTCATTGTTTGGGGCA | Fine mapping |
| TG-57 | 1 | CCACTAGTTGGAGCAGACAC | CGTCTTCCGGAAATTGTTGG | Fine mapping |

Fig. S2. Graphical representation of chromosome segment substitution lines (CSSLs). Each row means one of the CSSLs, with yellow filling indicating homozygosity for Zhonghui 9308, red filling indicating homozygosity for Xieqingzao B, and blue filling indicating heterozygosity.
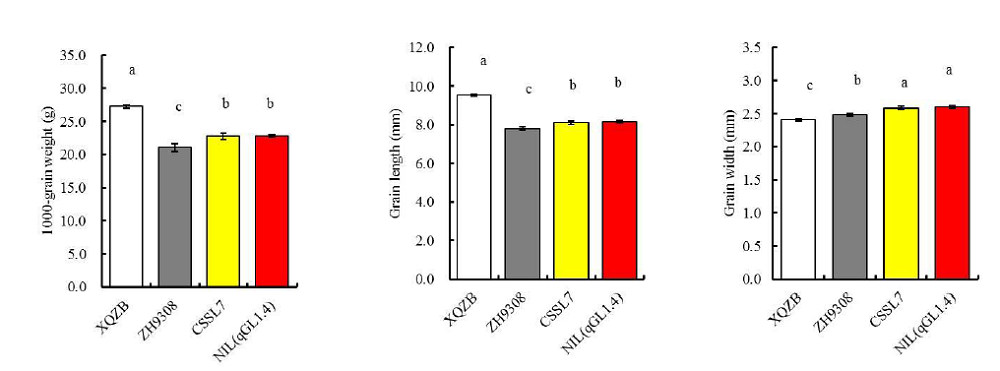
Fig. S3. Phenotypic comparisons among Xieqingzao B (XQZB), Zhonghui 9308 (ZH9308), CSSL7 and NIL(qGL1.4).Data represent Mean ± SE (n = 10). The same lowercase letters above the bars indicate there were no significant differences at (P < 0.05) according to Turkey’s t-test.
| QTL | Position | Marker | LOD | Add | PVE (%) |
|---|---|---|---|---|---|
| qTGW1 | Long arm of chromosome 1 | InDel15-RM12276 | 2.71 | 1.11 | 15.36 |
| qTGW6 | Short arm of chromosome 6 | InDell90-RM20069 | 2.15 | -1.28 | 10.48 |
Table S3. QTLs detected in CSSLs for 1000-grain weight.
| QTL | Position | Marker | LOD | Add | PVE (%) |
|---|---|---|---|---|---|
| qTGW1 | Long arm of chromosome 1 | InDel15-RM12276 | 2.71 | 1.11 | 15.36 |
| qTGW6 | Short arm of chromosome 6 | InDell90-RM20069 | 2.15 | -1.28 | 10.48 |
| Trait | 1000-grain weight | Grain length-width ratio | Grain length |
|---|---|---|---|
| Grain length-width ratio | 0.06 | ||
| Grain length | 0.68** | 0.29** | |
| Grain width | 0.49** | -0.65** | 0.50** |
Table S4. Correlation coefficient among grain size traits in F2 population.
| Trait | 1000-grain weight | Grain length-width ratio | Grain length |
|---|---|---|---|
| Grain length-width ratio | 0.06 | ||
| Grain length | 0.68** | 0.29** | |
| Grain width | 0.49** | -0.65** | 0.50** |
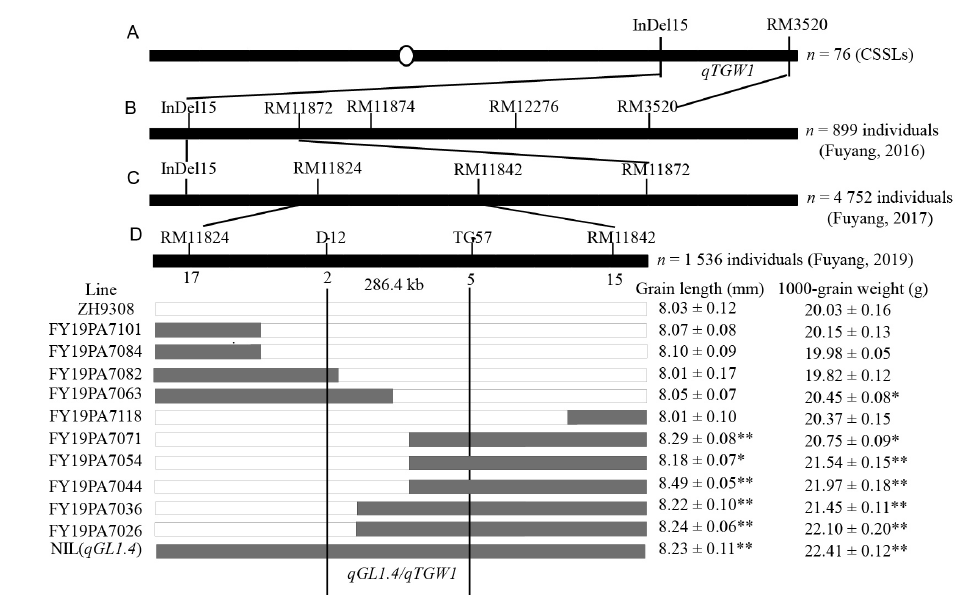
Fig. 1. Genetic mapping, dissection and physical maps of qGL1.4 on chromosome l.A, qGL1.4 was primarily mapped to the region between InDel15 and RM3520 on chromosome 1. CSSL, Chromosome segment substitution line. B, Validation of qGL1.4 based on the secondary F2 populations. The locus was mapped to the region between InDel15 and RM11872. C, Dissection of qGL1.4 based on the F2:3 populations and selection of homozygous recombinants. D, Fine mapping of qGL1.4. The allele was mapped to 286.4 kb between D-12 and TG-57 in F4:5 population. NIL, Near-isogenic line. * and ** indicate the significant differences at the 0.05 and 0.01 levels, respectively.
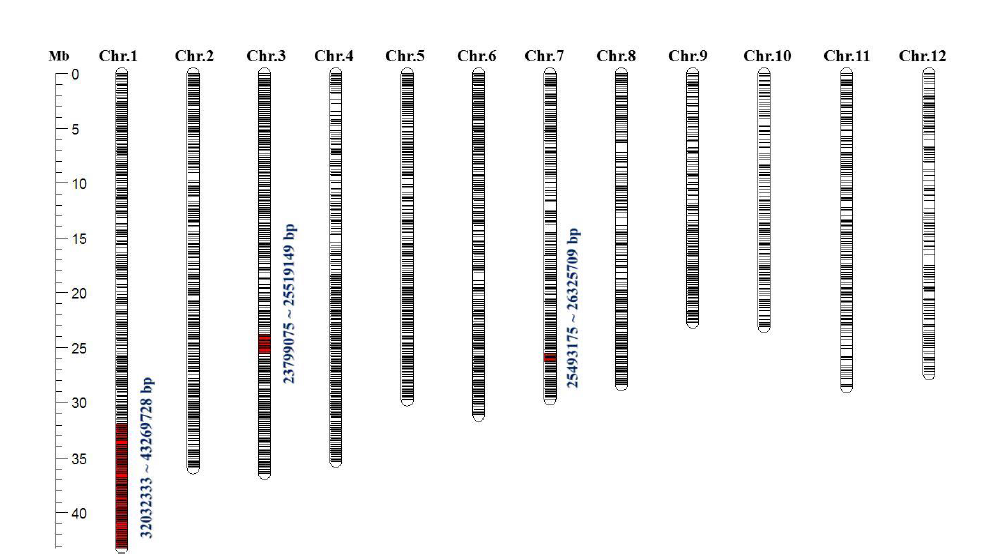
Fig. S6. Graphical genotype of CSSL7 derived from Xieqingzao B/Zhonghui 9308. The black lines represent all 6 782 SNPs across the 12 chromosomes. The red lines on chromosomes 1, 3 and 7 represent polymorphic SNPs in the introgression segments from Xieqingzao B. The physical positions of substituted segments are marked with blue numbers.
| [1] | Dong Q, Zhang Z H, Wang L L, Zhu Y J, Fan Y Y, Mou T M, Ma L Y, Zhuang J Y. 2018. Dissection and fine-mapping of two QTL for grain size linked in a 460-kb region on chromosome 1 of rice. Rice, 11(1): 44. |
| [2] | Fan C C, Yu S B, Wang C R, Xing Y Z. 2009. A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor Appl Genet, 118(3): 465-472. |
| [3] | Fitzgerald M A, McCouch S R, Hall R D. 2009. Not just a grain of rice: The quest for quality. Trends Plant Sci, 14(3): 133-139. |
| [4] | Li N, Xu R, Duan P G, Li Y H. 2018. Control of grain size in rice. Plant Reprod, 31(3): 237-251. |
| [5] | Madoka Y, Kashiwagi T, Hirotsu N, Ishimaru K. 2008. Indian rice ‘Kasalath’ contains genes that improve traits of Japanese premium rice ‘Koshihikari’. Theor Appl Genet, 116: 603-612. |
| [6] | Qi L, Ding Y B, Zheng X M, Xu R, Zhang L H, Wang Y Y, Wang X M, Zhang L F,·Cheng Y L, Qiao W H, Yang Q W. 2018. Fine mapping and identification of a novel locus qGL12. 2 control grain length in wild rice(Oryza rufipogon Griff.). Theor Appl Genet, 131(7): 1497-1508. |
| [7] | Qi L, Sun Y, Li J, Su L, Zheng X M, Wang X M, Li K M, Yang Q W, Qiao W H. 2017. Identify QTL for grain size and weight in common wild rice using chromosome segment substitution lines across six environments. Breeding Sci, 67: 472-482. |
| [8] | Ruan B P, Shang L G, Zhang B, Hu J, Wang Y X, Lin H, Zhang A P, Liu C L, Peng Y L, Zhu L, Ren D Y, Shen L, Dong G J, Zhang G H, Zeng D L, Guo L B, Qian Q, Gao Z Y. 2020. Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol, 227(2): 629-640. |
| [9] | Singh R, Singh A K, Sharma T R, Singh A, Singh N K. 2012. Fine mapping of grain length QTLs on chromosomes 1 and 7 in Basmati rice (Oryza sativa L.). J Plant Biochem Biotechnol, 21(2): 157-166. |
| [10] | Wang A, Hou Q Q, Si L H, Huang X H, Luo J H, Lu D F, Zhu J J, Shangguan Y Y, Miao J H, Xie Y F, Wang Y C, Zhao Q, Feng Q, Zhou C C, Li Y, Fan D L, Lu Y, Tian Q, Wang Z X, Han B. 2019a. The PLATZ transcription factorGL6 affects grain length and number in rice. Plant Physiol, 180(4): 2077-2090. |
| [11] | Wang W H, Wang L L, Zhu Y J, Fan Y Y, Zhuang J Y. 2019b. Fine-mapping of qTGW1.2a, a quantitative trait locus for 1000-grain weight in rice. Rice Sci, 26(4): 220-228. |
| [12] | Zhang B, Shang L G, Ruan B P, Zhang A P, Yang S L, Jiang H Z, Liu C L, Hong K, Lin H, Gao Z Y, Hu J, Zeng D, Guo L B, Qian Q. 2019. Development of three Sets of high-throughput genotyped rice chromosome segment substitution lines and QTL mapping for eleven traits. Rice, 12: 33. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||