
Rice Science ›› 2024, Vol. 31 ›› Issue (5): 507-525.DOI: 10.1016//j.rsci.2024.06.006
• Review • Previous Articles Next Articles
Liang Liang, Wang Chenchang, Chen Tao( )
)
Received:2024-04-04
Accepted:2024-06-05
Online:2024-09-28
Published:2024-10-11
Contact:
Chen Tao (chentao@hznu.edu.cn)
Liang Liang, Wang Chenchang, Chen Tao. Advances in Understanding Cadmium Stress and Breeding of Cadmium-Tolerant Crops[J]. Rice Science, 2024, 31(5): 507-525.
Add to citation manager EndNote|Ris|BibTeX
| Gene/Transporter | Function/Role | Reference |
|---|---|---|
| AtNRAMP3 | Cd absorption in Arabidopsis | Thomine et al, |
| PCR1, PCR2 | Cd transfer in Arabidopsis | Song et al, |
| OsIRT1, OsIRT2 | Cd influx transport in rice | Ishimaru et al, |
| ABCG36/PDR8 | Cd efflux in Arabidopsis | Kim et al, |
| OsHMA3 | Cd trafficking from rice roots to vacuoles | Ueno et al, |
| OsLCT1 | Cd transportation and accumulation in rice | Uraguchi et al, |
| OsNRAMP5 | Mn/Cd transport in rice | Takahashi et al, |
| OsHMA2 | Cd and Zn transport to aerial parts in rice | Yamaji et al, |
| OsZIP3 | Cd transport from roots to shoots in rice | Sasaki et al, |
| OsZIP6 | Cd transport from roots to shoots in rice | Kavitha et al, |
| OsZIP7 | Cd transport to developing tissues and grains in rice | Sasaki et al, |
| HvNRAMP5 | Cd uptake in barley | Wu et al, |
| OsNRAMP1 | Mn/Cd transport in rice | Tang et al, |
| CAL1 | Cd binding and secretion in rice | Luo et al, |
| OsCCX2 | Cd loading into xylem vessels in rice | Hao et al, |
| TpNRAMP5 | Cd concentration increase in Polish wheat | Peng et al, |
| OsABCG36 | Cd efflux pump in rice | Fu et al, |
| OsCd1 | Cd uptake in rice root and accumulation in grain | Yan et al, |
| OsZIP5, OsZIP9 | Cd root uptake in rice | Tan et al, |
Table 1. Key transporter genes involved in cadmium (Cd) stress response in various plant species.
| Gene/Transporter | Function/Role | Reference |
|---|---|---|
| AtNRAMP3 | Cd absorption in Arabidopsis | Thomine et al, |
| PCR1, PCR2 | Cd transfer in Arabidopsis | Song et al, |
| OsIRT1, OsIRT2 | Cd influx transport in rice | Ishimaru et al, |
| ABCG36/PDR8 | Cd efflux in Arabidopsis | Kim et al, |
| OsHMA3 | Cd trafficking from rice roots to vacuoles | Ueno et al, |
| OsLCT1 | Cd transportation and accumulation in rice | Uraguchi et al, |
| OsNRAMP5 | Mn/Cd transport in rice | Takahashi et al, |
| OsHMA2 | Cd and Zn transport to aerial parts in rice | Yamaji et al, |
| OsZIP3 | Cd transport from roots to shoots in rice | Sasaki et al, |
| OsZIP6 | Cd transport from roots to shoots in rice | Kavitha et al, |
| OsZIP7 | Cd transport to developing tissues and grains in rice | Sasaki et al, |
| HvNRAMP5 | Cd uptake in barley | Wu et al, |
| OsNRAMP1 | Mn/Cd transport in rice | Tang et al, |
| CAL1 | Cd binding and secretion in rice | Luo et al, |
| OsCCX2 | Cd loading into xylem vessels in rice | Hao et al, |
| TpNRAMP5 | Cd concentration increase in Polish wheat | Peng et al, |
| OsABCG36 | Cd efflux pump in rice | Fu et al, |
| OsCd1 | Cd uptake in rice root and accumulation in grain | Yan et al, |
| OsZIP5, OsZIP9 | Cd root uptake in rice | Tan et al, |
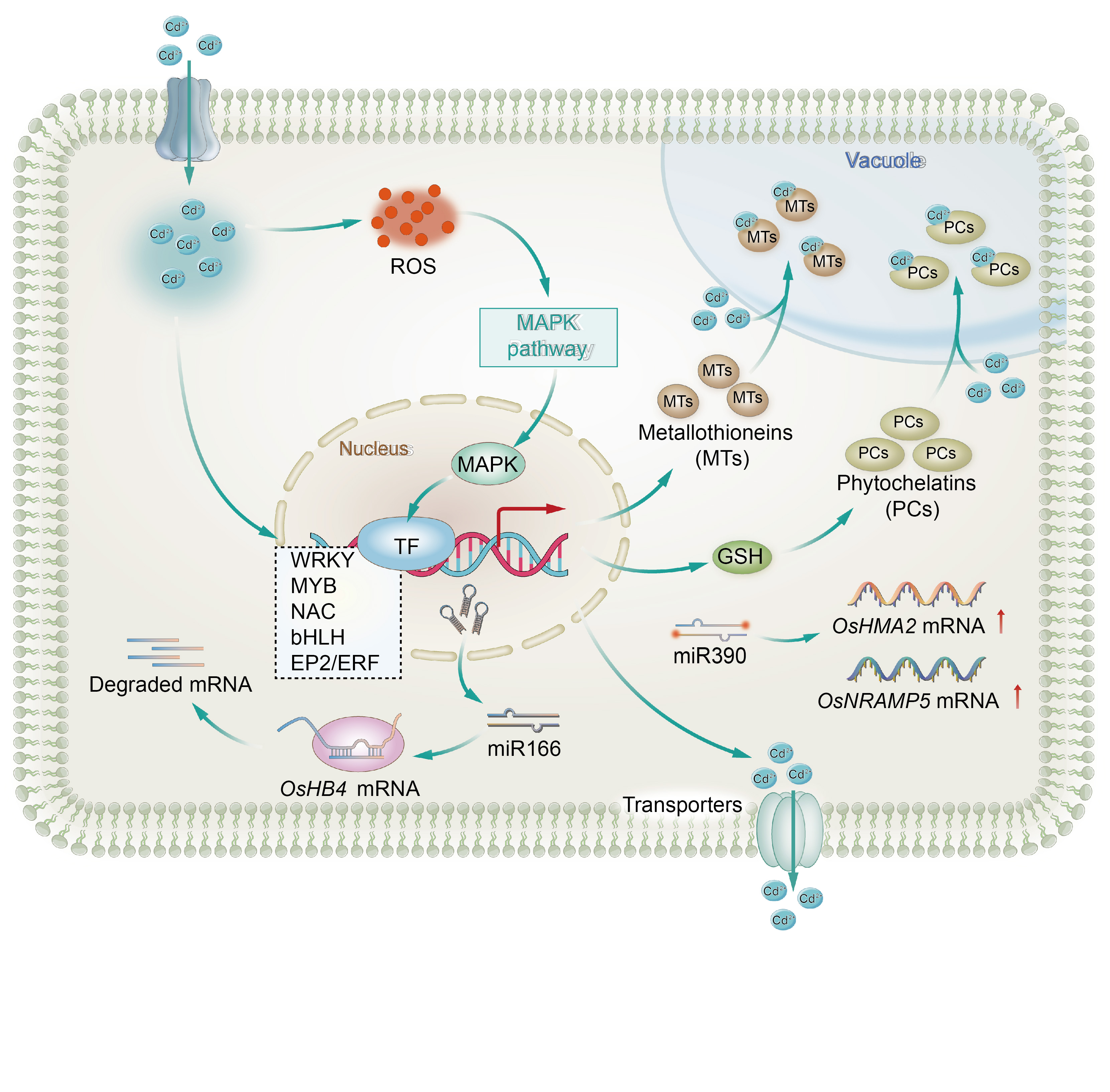
Fig. 1. Presumed transcriptional network associated with cadmium (Cd) signaling in rice. Upon entry of Cd2+ (blue circles) into the cell, it leads to the accumulation of reactive oxygen species (ROS, red ellipsoids), activing the mitogen-activated protein kinase pathway (MAPK, green ellipsoids) that transmits the signal to the nucleus. This influences transcription factors (TF, blue ellipsoids), which, in turn, regulate the downstream expression of Cd transporters (aggregate of ellipsoids), metal-binding proteins (brown ellipsoids), and metal chelators (yellow ellipsoids). Additionally, miR390 increases the expression of OsNRAMP5 and OsHMA2, enhancing Cd uptake and transport in rice, while miR166 degrades OsHB4 mRNA, reducing OsHB4 expression and thereby decreasing Cd accumulation and increasing Cd tolerance. GSH, Glutathione.
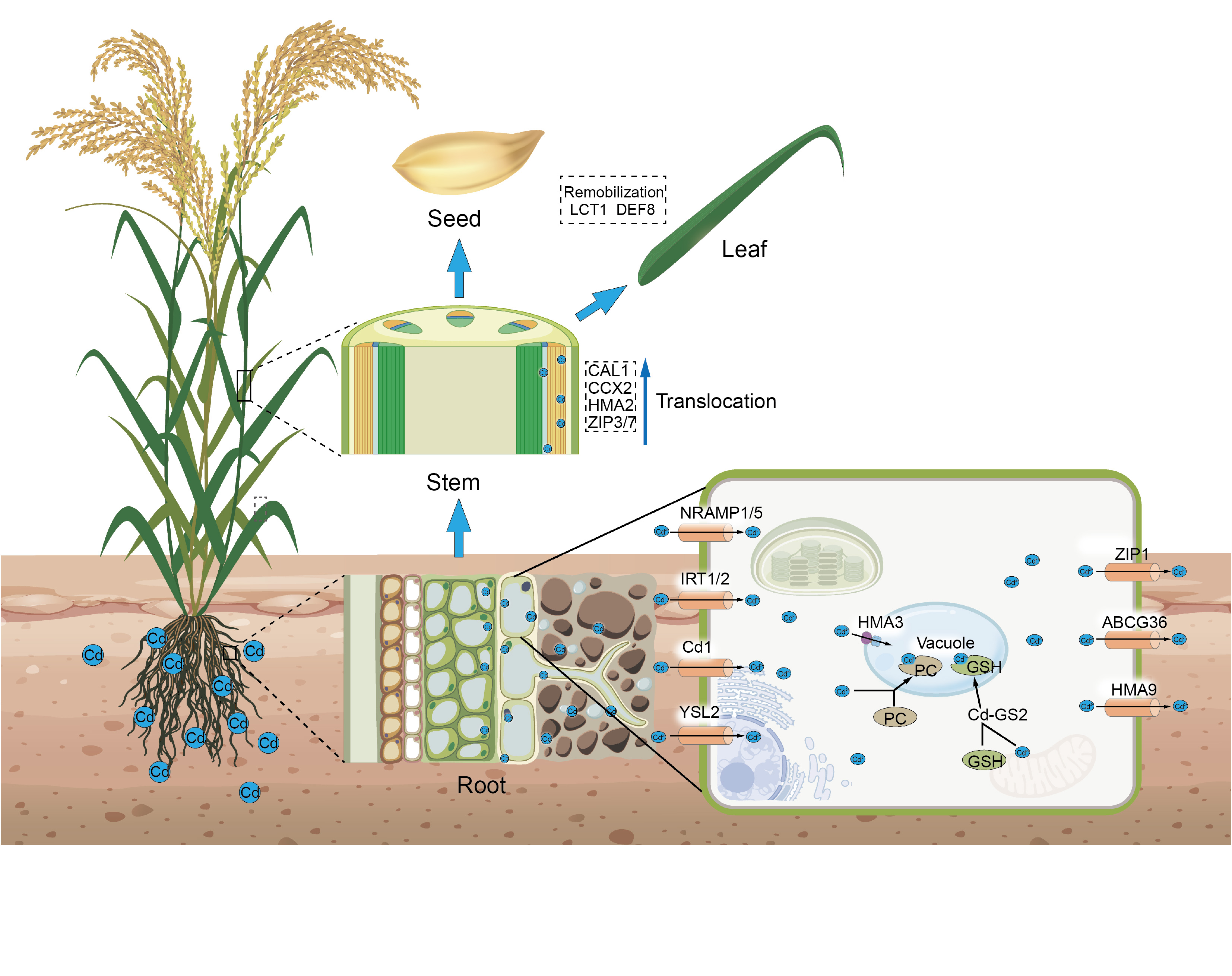
Fig. 2. Schematic diagram of cadmium (Cd) uptake, transport, and remobilization in rice. The blue dots represent Cd and Cd2+, while the cylindrical shapes on the cell membrane depict proteins that facilitate the entry and exit of Cd2+. The ellipsoids, colored brown and green, represent the chelators of phosphatidylcholines (PCs) and glutathione (GSH), respectively. Proteins within the dashed rectangular structures are involved in the transport of Cd in the stem, as well as those participating in the mobilization of Cd in seeds and leaves.
| [1] | Abbas T, Rizwan M, Ali S, Adrees M, Zia-ur-Rehman M, Qayyum M F, Ok Y S, Murtaza G. 2018. Effect of biochar on alleviation of cadmium toxicity in wheat (Triticum aestivum L.) grown on Cd-contaminated saline soil. Environ Sci Pollut Res, 25(26): 25668-25680. |
| [2] | Ahmad I, Naeem M, Khan N A, Samiullah. 2009. Effects of cadmium stress upon activities of antioxidative enzymes, photosynthetic rate, and production of phytochelatins in leaves and chloroplasts of wheat cultivars differing in yield potential. Photosynthetica, 47(1): 146-151. |
| [3] | Ai H, Wu D X, Li C L, Hou M M. 2022. Advances in molecular mechanisms underlying cadmium uptake and translocation in rice. Front Plant Sci, 13: 1003953. |
| [4] | Alsaleh A, Baloch F S, Sesiz U, Nadeem M A, Hatipoğlu R, Erbakan M, Özkan H. 2022. Marker-assisted selection and validation of DNA markers associated with cadmium content in durum wheat germplasm. Crop Pasture Sci, 73(8): 943-956. |
| [5] | Angulo-Bejarano P I, Puente-Rivera J, Cruz-Ortega R. 2021. Metal and metalloid toxicity in plants: An overview on molecular aspects. Plants, 10(4): 635. |
| [6] | Azevedo H, Glória Pinto C G, Santos C. 2005. Cadmium effects in sunflower: Membrane permeability and changes in catalase and peroxidase activity in leaves and calluses. J Plant Nutr, 28(12): 2233-2241. |
| [7] |
Balestrasse K B, Benavides M P, Gallego S M, Tomaro M L. 2003. Effect of cadmium stress on nitrogen metabolism in nodules and roots of soybean plants. Funct Plant Biol, 30(1): 57-64.
DOI PMID |
| [8] | Baryla A, Carrier P, Franck F, Coulomb C, Sahut C, Havaux M. 2001. Leaf chlorosis in oilseed rape plants (Brassica napus) grown on cadmium-polluted soil: Causes and consequences for photosynthesis and growth. Planta, 212(5/6): 696-709. |
| [9] |
Benitez E R, Hajika M, Takahashi R. 2012. Single-base substitution in P1B-ATPase gene is associated with a major QTL for seed cadmium concentration in soybean. J Hered, 103(2): 278-286.
DOI PMID |
| [10] | Brunetti P, Zanella L, de Paolis A, di Litta D, Cecchetti V, Falasca G, Barbieri M, Altamura M M, Costantino P, Cardarelli M. 2015. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J Exp Bot, 66(13): 3815-3829. |
| [11] | Cao X, Wang H T, Zhuang D F, Zhu H, Du Y L, Cheng Z B, Cui W N, Rogers H J, Zhang Q R, Jia C J, Yang Y S, Tai P D, Xie F T, Liu W. 2018. Roles of MSH2 and MSH6 in cadmium-induced G2/M checkpoint arrest in Arabidopsis roots. Chemosphere, 201: 586-594. |
| [12] | Cao Z Z, Qin M L, Lin X Y, Zhu Z W, Chen M X. 2018. Sulfur supply reduces cadmium uptake and translocation in rice grains (Oryza sativa L.) by enhancing iron plaque formation, cadmium chelation and vacuolar sequestration. Environ Pollut, 238: 76-84. |
| [13] | Cao Z Z, Lin X Y, Yang Y J, Guan M Y, Xu P, Chen M X. 2019. Gene identification and transcriptome analysis of low cadmium accumulation rice mutant (lcd1) in response to cadmium stress using MutMap and RNA-seq. BMC Plant Biol, 19(1): 250. |
| [14] |
Chen C H, Zhou Q X, Cai Z. 2014. Effect of soil HHCB on cadmium accumulation and phytotoxicity in wheat seedlings. Ecotoxicology, 23(10): 1996-2004.
DOI PMID |
| [15] | Chen G, Du R Y, Wang X. 2023. Genetic regulation mechanism of cadmium accumulation and its utilization in rice breeding. Int J Mol Sci, 24(2): 1247. |
| [16] |
Ci D W, Jiang D, Dai T B, Jing Q, Cao W X. 2009. Effects of cadmium on plant growth and physiological traits in contrast wheat recombinant inbred lines differing in cadmium tolerance. Chemosphere, 77(11): 1620-1625.
DOI PMID |
| [17] | Ci D W, Jiang D, Wollenweber B, Dai T B, Jing Q, Cao W X. 2010. Cadmium stress in wheat seedlings: Growth, cadmium accumulation and photosynthesis. Acta Physiol Plant, 32(2): 365-373. |
| [18] |
Clemens S. 2006. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie, 88(11): 1707-1719.
DOI PMID |
| [19] |
Cobbett C, Goldsbrough P. 2002. Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol, 53: 159-182.
PMID |
| [20] | Considine M J, Foyer C H. 2021. Oxygen and reactive oxygen species-dependent regulation of plant growth and development. Plant Physiol, 186(1): 79-92. |
| [21] |
DalCorso G, Farinati S, Furini A. 2010. Regulatory networks of cadmium stress in plants. Plant Signal Behav, 5(6): 663-667.
DOI PMID |
| [22] | Das N, Bhattacharya S, Bhattacharyya S, Maiti M K. 2017. Identification of alternatively spliced transcripts of rice phytochelatin synthase 2 gene OsPCS2 involved in mitigation of cadmium and arsenic stresses. Plant Mol Biol, 94(1/2): 167-183. |
| [23] | Dheri G S, Brar M S, Malhi S S. 2007. Influence of phosphorus application on growth and cadmium uptake of spinach in two cadmium-contaminated soils. J Plant Nutr Soil Sci, 170(4): 495-499. |
| [24] | di Toppi S L, Gabbrielli R. 1999. Response to cadmium in higher plants. Environ Exp Bot, 41(2): 105-130. |
| [25] |
Ding Y F, Ye Y Y, Jiang Z H, Wang Y, Zhu C. 2016. MicroRNA390 is involved in cadmium tolerance and accumulation in rice. Front Plant Sci, 7: 235.
DOI PMID |
| [26] |
Ding Y F, Gong S H, Wang Y, Wang F J, Bao H, Sun J W, Cai C, Yi K K, Chen Z X, Zhu C. 2018. MicroRNA166 modulates cadmium tolerance and accumulation in rice. Plant Physiol, 177(4): 1691-1703.
DOI PMID |
| [27] | Ehsan S, Ali S, Noureen S, Mahmood K, Farid M, Ishaque W, Shakoor M B, Rizwan M. 2014. Citric acid assisted phytoremediation of cadmium by Brassica napus L. Ecotoxicol Environ Saf, 106: 164-172. |
| [28] | Eker S, Erdem H, Yazici M A, Barut H, Heybet E H. 2013. Effects of cadmium on growth and nutrient composition of bread and durum wheat genotypes. Fresenius Environ Bull, 22(6): 1779-1786. |
| [29] | Eriksson J E. 1989. The influence of pH, soil type and time on adsorbtion and uptake by plants of Cd added to the soil. Water Air Soil Pollut, 48(3): 317-335. |
| [30] | Farooq M A, Ali S, Hameed A, Bharwana S A, Rizwan M, Ishaque W, Farid M, Mahmood K, Iqbal Z. 2016. Cadmium stress in cotton seedlings: Physiological, photosynthesis and oxidative damages alleviated by glycinebetaine. S Afr N J Bot, 104: 61-68. |
| [31] |
Freeman J L, Persans M W, Nieman K, Albrecht C, Peer W, Pickering I J, Salt D E. 2004. Increased glutathione biosynthesis plays a role in nickel tolerance in thlaspi nickel hyperaccumulators. Plant Cell, 16(8): 2176-2191.
DOI PMID |
| [32] | Fodor E, Szabó-Nagy A, Erdei L. 1995. The effects of cadmium on the fluidity and H+-ATPase activity of plasma membrane from sunflower and wheat roots. J Plant Physiol, 147(1): 87-92. |
| [33] |
Fu S, Lu Y S, Zhang X, Yang G Z, Chao D, Wang Z G, Shi M X, Chen J G, Chao D Y, Li R B, Ma J F, Xia J X. 2019. The ABC transporter ABCG36 is required for cadmium tolerance in rice. J Exp Bot, 70(20): 5909-5918.
DOI PMID |
| [34] | Ge Y, Hendershot W. 2005. Modeling sorption of Cd, Hg and Pb in soils by the NICA-donnan model. Soil Sediment Contam, 14(1): 53-69. |
| [35] | Genchi G, Sinicropi M S, Lauria G, Carocci A, Catalano A. 2020. The effects of cadmium toxicity. Int J Environ Res Public Health, 17(11): 3782. |
| [36] | Gu Q, Wang C Y, Xiao Q Q, Chen Z P, Han Y. 2021. Melatonin confers plant cadmium tolerance: An update. Int J Mol Sci, 22(21): 11704. |
| [37] | Guo T R, Zhang G P, Zhou M X, Wu F B, Chen J X. 2007. Influence of aluminum and cadmium stresses on mineral nutrition and root exudates in two barley cultivars. Pedosphere, 17(4): 505-512. |
| [38] | Haider F U, Cai L Q, Coulter J A, Cheema S A, Wu J, Zhang R Z, Ma W J, Farooq M. 2021. Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicol Environ Saf, 211: 111887. |
| [39] | Hamid Y, Tang L, Sohail M I, Cao X R, Hussain B, Aziz M Z, Usman M, He Z L, Yang X E. 2019. An explanation of soil amendments to reduce cadmium phytoavailability and transfer to food chain. Sci Total Environ, 660: 80-96. |
| [40] |
Hao X H, Zeng M, Wang J, Zeng Z W, Dai J L, Xie Z J, Yang Y Z, Tian L F, Chen L B, Li D P. 2018. A node-expressed transporter OsCCX2 is involved in grain cadmium accumulation of rice. Front Plant Sci, 9: 476.
DOI PMID |
| [41] |
Hasan S A, Fariduddin Q, Ali B, Hayat S, Ahmad A. 2009. Cadmium: Toxicity and tolerance in plants. J Environ Biol, 30(2): 165-174.
PMID |
| [42] | Hassinen V H, Tervahauta A I, Schat H, Kärenlampi S O. 2011. Plant metallothioneins: Metal chelators with ROS scavenging activity? Plant Biol, 13(2): 225-232. |
| [43] | He L L, Ma X L, Li Z Z, Jiao Z L, Li Y Q, Ow D W. 2016. Maize OXIDATIVE STRESS2 homologs enhance cadmium tolerance in Arabidopsis through activation of a putative SAM-dependent methyltransferase gene. Plant Physiol, 171(3): 1675-1685. |
| [44] | He X L, Fan S K, Zhu J, Guan M Y, Liu X X, Zhang Y S, Jin C W. 2017. Iron supply prevents Cd uptake in Arabidopsis by inhibiting IRT1 expression and favoring competition between Fe and Cd uptake. Plant Soil, 416(1): 453-462. |
| [45] | Heiss S, Wachter A, Bogs J, Cobbett C, Rausch T. 2003. Phytochelatin synthase (PCS) protein is induced in Brassica juncea leaves after prolonged Cd exposure. J Exp Bot, 54(389): 1833-1839. |
| [46] |
Heyno E, Klose C, Krieger-Liszkay A. 2008. Origin of cadmium- induced reactive oxygen species production: Mitochondrial electron transfer versus plasma membrane NADPH oxidase. New Phytol, 179(3): 687-699.
DOI PMID |
| [47] |
Hua K, Zhang J S, Botella J R, Ma C L, Kong F J, Liu B H, Zhu J K. 2019. Perspectives on the application of genome-editing technologies in crop breeding. Mol Plant, 12(8): 1047-1059.
DOI PMID |
| [48] | Huybrechts M, Cuypers A, Deckers J, Iven V, Vandionant S, Jozefczak M, Hendrix S. 2019. Cadmium and plant development: An agony from seed to seed. Int J Mol Sci, 20(16): 3971. |
| [49] | Ishikawa S. 2020. Mechanisms of cadmium accumulation in rice grains and molecular breeding for its reduction. Soil Sci Plant Nutr, 66(1): 28-33. |
| [50] |
Ishikawa S, Ishimaru Y, Igura M, Kuramata M, Abe T, Senoura T, Hase Y, Arao T, Nishizawa N K, Nakanishi H. 2012. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc Natl Acad Sci USA, 109(47): 19166-19171.
DOI PMID |
| [51] |
Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa N K. 2006. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J, 45(3): 335-346.
DOI PMID |
| [52] | Islam M M, Hoque M A, Okuma E, Banu M N A, Shimoishi Y, Nakamura Y, Murata Y. 2009. Exogenous proline and glycinebetaine increase antioxidant enzyme activities and confer tolerance to cadmium stress in cultured tobacco cells. J Plant Physiol, 166(15): 1587-1597. |
| [53] | Järup L. 2003. Hazards of heavy metal contamination. Br Med Bull, 68: 167-182. |
| [54] | Jia J, Bai J H, Xiao R, Tian S M, Wang D W, Wang W, Zhang G L, Cui H, Zhao Q Q. 2022. Fractionation, source, and ecological risk assessment of heavy metals in cropland soils across a 100-year reclamation chronosequence in an estuary, South China. Sci Total Environ, 807: 151725. |
| [55] | Jiang G L. 2013. Molecular markers and marker-assisted breeding in plants. Plant Breed Lab Fields, 3: 45-83. |
| [56] | Jin C W, Mao Q Q, Luo B F, Lin X Y, Du S T. 2013. Mutation of mpk6 enhances cadmium tolerance in Arabidopsis plants by alleviating oxidative stress. Plant Soil, 371(1): 387-396. |
| [57] | Kalai T, Bouthour D, Manai J, Ben Kaab L B, Gouia H. 2016. Salicylic acid alleviates the toxicity of cadmium on seedling growth, amylases and phosphatases activity in germinating barley seeds. Arch Agron Soil Sci, 62(6): 892-904. |
| [58] | Kavitha P G, Kuruvilla S, Mathew M K. 2015. Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.). Plant Physiol Biochem, 97: 165-174. |
| [59] | Khan N A, Samiullah, Singh S, Nazar R. 2007. Activities of antioxidative enzymes, sulphur assimilation, photosynthetic activity and growth of wheat (Triticum aestivum) cultivars differing in yield potential under cadmium stress. J Agron Crop Sci, 193(6): 435-444. |
| [60] | Kim D Y, Bovet L, Maeshima M, Martinoia E, Lee Y. 2007. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J, 50(2): 207-218. |
| [61] | Kim K R, Owens G, Naidu R. 2009. Heavy metal distribution, bioaccessibility, and phytoavailability in long-term contaminated soils from Lake Macquarie, Australia. Aust J Soil Res, 47(2): 166-176. |
| [62] |
Krämer U, Talke I N, Hanikenne M. 2007. Transition metal transport. FEBS Lett, 581(12): 2263-2272.
DOI PMID |
| [63] | Krantev A, Yordanova R, Janda T, Szalai G, Popova L. 2008. Treatment with salicylic acid decreases the effect of cadmium on photosynthesis in maize plants. J Plant Physiol, 165(9): 920-931. |
| [64] |
Küpper H, Kochian L V. 2010. Transcriptional regulation of metal transport genes and mineral nutrition during acclimatization to cadmium and zinc in the Cd/Zn hyperaccumulator, Thlaspi caerulescens (Ganges population). New Phytol, 185(1): 114-129.
DOI PMID |
| [65] | la Rocca N, Andreoli C, Giacometti G M, Rascio N, Moro I. 2009. Responses of the Antarctic microalga Koliella antarctica (Trebouxiophyceae, Chlorophyta) to cadmium contamination. Photosynthetica, 47(3): 471-479. |
| [66] | Landberg T, Greger M. 2003. Influence of N and N supplementation on Cd accumulation in wheat grains. In: Proceedings of the 7th International Conference on the Biogeochemistry of Trace Elements. 15-19 June 2003. SLU Service Upsala, Swiss. |
| [67] |
Lee S, Kim Y Y, Lee Y, An G. 2007. Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol, 145(3): 831-842.
DOI PMID |
| [68] |
Lei G J, Fujii-Kashino M, Wu D Z, Hisano H, Saisho D, Deng F L, Yamaji N, Sato K, Zhao F J, Ma J F. 2020. Breeding for low cadmium barley by introgression of a Sukkula-like transposable element. Nat Food, 1(8): 489-499.
DOI PMID |
| [69] |
Lin T T, Yang W N, Lu W, Wang Y, Qi X T. 2017. Transcription factors PvERF15 and PvMTF-1 form a cadmium stress transcriptional pathway. Plant Physiol, 173(3): 1565-1573.
DOI PMID |
| [70] | Liu K, Lv J L, He W X, Zhang H, Cao Y F, Dai Y C. 2015. Major factors influencing cadmium uptake from the soil into wheat plants. Ecotoxicol Environ Saf, 113: 207-213. |
| [71] | Liu S M, Jiang J, Liu Y, Meng J, Xu S L, Tan Y Y, Li Y F, Shu Q Y, Huang J Z. 2019. Characterization and evaluation of OsLCT1 and OsNramp5 mutants generated through CRISPR/Cas9-mediated mutagenesis for breeding low Cd rice. Rice Sci, 26(2): 88-97. |
| [72] | Liu X L, Zhang S Z, Shan X Q, Christie P. 2007. Combined toxicity of cadmium and arsenate to wheat seedlings and plant uptake and antioxidative enzyme responses to cadmium and arsenate co-contamination. Ecotoxicol Environ Saf, 68(2): 305-313. |
| [73] | Liu X S, Feng S J, Zhang B Q, Wang M Q, Cao H W, Rono J K, Chen X, Yang Z M. 2019. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol, 19(1): 283. |
| [74] | Luo J S, Huang J, Zeng D L, Peng J S, Zhang G B, Ma H L, Guan Y, Yi H Y, Fu Y L, Han B, Lin H X, Qian Q, Gong J M. 2018. A defensin-like protein drives cadmium efflux and allocation in rice. Nat Commun, 9(1): 645. |
| [75] |
Lux A, Martinka M, Vaculík M, White P J. 2011. Root responses to cadmium in the rhizosphere: A review. J Exp Bot, 62(1): 21-37.
DOI PMID |
| [76] | Lv Q M, Li W G, Sun Z Z, Ouyang N, Jing X, He Q, Wu J, Zheng J K, Zheng J T, Tang S Q, Zhu R S, Tian Y, Duan M J, Tan Y N, Yu D, Sheng X B, Sun X W, Jia G F, Gao H Z, Zeng Q, Li Y F, Tang L, Xu Q S, Zhao B R, Huang Z Y, Lu H F, Li N, Zhao J, Zhu L H, Li D, Yuan L P, Yuan D Y. 2020. Resequencing of 1,143 indica rice accessions reveals important genetic variations and different heterosis patterns. Nat Commun, 11: 4778. |
| [77] | Malan H L, Farrant J M. 1998. Effects of the metal pollutants cadmium and nickel on soybean seed development. Seed Sci Res, 8(4): 445-453. |
| [78] | Mao Q Q, Guan M Y, Lu K X, Du S T, Fan S K, Ye Y Q, Lin X Y, Jin C W. 2014. Inhibition of nitrate transporter 1.1-controlled nitrate uptake reduces cadmium uptake in Arabidopsis. Plant Physiol, 166(2): 934-944. |
| [79] | Markovska Y K, Gorinova N I, Nedkovska M P, Miteva K M. 2009. Cadmium-induced oxidative damage and antioxidant responses in Brassica juncea plants. Biol Plant, 53(1): 151-154. |
| [80] |
Matusik J, Bajda T, Manecki M. 2008. Immobilization of aqueous cadmium by addition of phosphates. J Hazard Mater, 152(3): 1332-1339.
PMID |
| [81] | May M J, Vernoux T, Leaver C, van Montagu M, Inzé D. 1998. Glutathione homeostasis in plants: Implications for environmental sensing and plant development. J Exp Bot, 49(321): 649-667. |
| [82] | Mills R F, Peaston K A, Runions J, Williams L E. 2012. HvHMA2, a P1B-ATPase from barley, is highly conserved among cereals and functions in Zn and Cd transport. PLoS One, 7(8): e42640. |
| [83] |
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. 2004. Reactive oxygen gene network of plants. Trends Plant Sci, 9(10): 490-498.
DOI PMID |
| [84] |
Miyadate H, Adachi S, Hiraizumi A, Tezuka K, Nakazawa N, Kawamoto T, Katou K, Kodama I, Sakurai K, Takahashi H, Satoh-Nagasawa N, Watanabe A, Fujimura T, Akagi H. 2011. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol, 189(1): 190-199.
DOI PMID |
| [85] | Nakanishi H, Ogawa I, Ishimaru Y, Mori S, Nishizawa N K. 2006. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci Plant Nutr, 52(4): 464-469. |
| [86] |
Nelson M T. 1986. Interactions of divalent cations with single calcium channels from rat brain synaptosomes. J Gen Physiol, 87(2): 201-222.
DOI PMID |
| [87] | Noor I, Sohail H, Sun J X, Nawaz M A, Li G H, Hasanuzzaman M, Liu J W. 2022. Heavy metal and metalloid toxicity in horticultural plants: Tolerance mechanism and remediation strategies. Chemosphere, 303: 135196. |
| [88] | Noor W, Umar S, Mir M Y, Shah D, Majeed G, Hafeez S, Yaqoob S, Gulzar A, Kamili A N. 2018. Effect of cadmium on growth, photosynthesis and nitrogen metabolism of crop plants. J Res Dev, 18: 100-108. |
| [89] |
Ortiz D F, Ruscitti T, McCue K F, Ow D W. 1995. Transport of metal-binding peptides by HMT1, a fission yeast ABC-type vacuolar membrane protein. J Biol Chem, 270(9): 4721-4728.
DOI PMID |
| [90] | Paradiso A, Berardino R, de Pinto M C, Sanità di Toppi L, Storelli M M, Tommasi F, De Gara L. 2008. Increase in ascorbate- glutathione metabolism as local and precocious systemic responses induced by cadmium in durum wheat plants. Plant Cell Physiol, 49(3): 362-374. |
| [91] | Parekh D, Puranik R M, Srivastava H S. 1990. Inhibition of chlorophyll biosynthesis by cadmium in greening maize leaf segments. Biochem Physiol Pflanz, 186(4): 239-242. |
| [92] | Park J, Song W Y, Ko D, Eom Y, Hansen T H, Schiller M, Lee T G, Martinoia E, Lee Y. 2012. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J, 69(2): 278-288. |
| [93] |
Peng F, Wang C, Zhu J S, Zeng J, Kang H Y, Fan X, Sha L N, Zhang H Q, Zhou Y H, Wang Y. 2018. Expression of TpNRAMP5, a metal transporter from Polish wheat (Triticum polonicum L.), enhances the accumulation of Cd, Co and Mn in transgenic Arabidopsis plants. Planta, 247(6): 1395-1406.
DOI PMID |
| [94] |
Penner G A, Bezte L J, Leisle D, Clarke J. 1995. Identification of RAPD markers linked to a gene governing cadmium uptake in durum wheat. Genome, 38(3): 543-547.
PMID |
| [95] | Puschenreiter M, Horak O, Friesl W, Hartl W. 2005. Low-cost agricultural measures to reduce heavy metal transfer into the food chain: A review. Plant Soil Environ, 51(1): 1-11. |
| [96] |
Qiao K, Gong L, Tian Y B, Wang H, Chai T Y. 2018. The metal- binding domain of wheat heavy metal ATPase 2 (TaHMA2) is involved in zinc/cadmium tolerance and translocation in Arabidopsis. Plant Cell Rep, 37(9): 1343-1352.
DOI PMID |
| [97] | Rahman A, Nahar K, Hasanuzzaman M, Fujita M. 2016. Manganese- induced cadmium stress tolerance in rice seedlings: Coordinated action of antioxidant defense, glyoxalase system and nutrient homeostasis. C R Biol, 339(11/12): 462-474. |
| [98] | Reddy C S, Cho M, Kaul T, Joeng J T, Kim K M. 2023. Pseudomonas fluorescens imparts cadmium stress tolerance in Arabidopsis thaliana via induction of AtPCR2 gene expression. J Genet Eng Biotechnol, 21(1): 8. |
| [99] |
Ribaut J M, de Vicente M C, Delannay X. 2010. Molecular breeding in developing countries: Challenges and perspectives. Curr Opin Plant Biol, 13(2): 213-218.
DOI PMID |
| [100] | Rizwan M, Meunier J D, Miche H, Keller C. 2012. Effect of silicon on reducing cadmium toxicity in durum wheat (Triticum turgidum L. cv. Claudio W.) grown in a soil with aged contamination. J Hazard Mater, 209/210: 326-334. |
| [101] | Rizwan M, Ali S, Adrees M, Rizvi H, Zia-Ur-Rehman M, Hannan F, Qayyum M F, Hafeez F, Ok Y S. 2016. Cadmium stress in rice: Toxic effects, tolerance mechanisms, and management: A critical review. Environ Sci Pollut Res Int, 23(18): 17859-17879. |
| [102] | Rochaix J D. 2011. Regulation of photosynthetic electron transport. Biochim Biophys Acta, 1807(3): 375-383. |
| [103] |
Romero-Puertas M C, Terrón-Camero L C, Peláez-Vico M Á, Olmedilla A, Sandalio L M. 2019. Reactive oxygen and nitrogen species as key indicators of plant responses to Cd stress. Environ Exp Bot, 161: 107-119.
DOI |
| [104] | Rossini F P, Martins J P R, Moreira S W, Conde L T, Clairvil E, da Conceição de Souza Braga P, Falqueto A R, Gontijo A B P L. 2022. In vitro morphophysiological responses of Alternanthera tenella colla (Amaranthaceae) to stress induced by cadmium and the attenuating action of silicon. Plant Cell Tissue Organ Cult, 150(1): 223-236. |
| [105] | Salsman E, Kumar A, AbuHammad W, Abbasabadi A O, Dobrydina M, Chao S, Li X H, Manthey F A, Elias E M. 2018. Development and validation of molecular markers for grain cadmium in durum wheat. Mol Breed, 38(3): 28. |
| [106] |
Sandalio L M, Dalurzo H C, Gómez M, Romero-Puertas M C, del Río L A. 2001. Cadmium-induced changes in the growth and oxidative metabolism of pea plants. J Exp Bot, 52: 2115-2126.
DOI PMID |
| [107] | Sarwar N, Saifullah, Malhi S S, Zia M H, Naeem A, Bibi S, Farid G. 2010. Role of mineral nutrition in minimizing cadmium accumulation by plants. J Sci Food Agric, 90(6): 925-937. |
| [108] | Sasaki A, Yamaji N, Mitani-Ueno N, Kashino M, Ma J F. 2015. A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J, 84(2): 374-384. |
| [109] |
Satoh-Nagasawa N, Mori M, Nakazawa N, Kawamoto T, Nagato Y, Sakurai K, Takahashi H, Watanabe A, Akagi H. 2012. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol, 53(1): 213-224.
DOI PMID |
| [110] |
Saxena I, Srikanth S, Chen Z. 2016. Cross talk between H2O2 and interacting signal molecules under plant stress response. Front Plant Sci, 7: 570.
DOI PMID |
| [111] |
Schützendübel A, Schwanz P, Teichmann T, Gross K, Langenfeld- Heyser R, Godbold D L, Polle A. 2001. Cadmium-induced changes in antioxidative systems, hydrogen peroxide content, and differentiation in Scots pine roots. Plant Physiol, 127(3): 887-898.
PMID |
| [112] | Shao G S, Chen M X, Wang D Y, Xu C M, Mou R X, Cao Z Y, Zhang X F. 2008. Using iron fertilizer to control Cd accumulation in rice plants: A new promising technology. Sci China C Life Sci, 51(3): 245-253. |
| [113] | Sheng Y B, Yan X X, Huang Y, Han Y Y, Zhang C, Ren Y B, Fan T T, Xiao F M, Liu Y S, Cao S Q. 2019. The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ, 42(3): 891-903. |
| [114] | Shim D, Hwang J U, Lee J, Lee S, Choi Y, An G, Martinoia E, Lee Y. 2009. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell, 21(12): 4031-4043. |
| [115] |
Shimo H, Ishimaru Y, An G, Yamakawa T, Nakanishi H, Nishizawa N K. 2011. Low cadmium (LCD), a novel gene related to cadmium tolerance and accumulation in rice. J Exp Bot, 62(15): 5727-5734.
DOI PMID |
| [116] | Somashekaraiah B V, Padmaja K, Prasad A R K. 1992. Phytotoxicity of cadmium ions on germinating seedlings of mung bean (Phaseolus vulgaris): Involvement of lipid peroxides in chlorphyll degradation. Physiol Plant, 85(1): 85-89. |
| [117] | Song W Y, Martinoia E, Lee J, Kim D, Kim D Y, Vogt E, Shim D, Choi K S, Hwang I, Lee Y. 2004. A novel family of Cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol, 135(2): 1027-1039. |
| [118] |
Sun N, Liu M, Zhang W T, Yang W N, Bei X J, Ma H, Qiao F, Qi X T. 2015. Bean metal-responsive element-binding transcription factor confers cadmium resistance in tobacco. Plant Physiol, 167(3): 1136-1148.
DOI PMID |
| [119] |
Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa N K. 2011. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J Exp Bot, 62(14): 4843-4850.
DOI PMID |
| [120] | Takahashi R, Ishimaru Y, Shimo H, Ogo Y, Senoura T, Nishizawa N K, Nakanishi H. 2012. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ, 35(11): 1948-1957. |
| [121] | Takahashi R, Ito M, Katou K, Sato K, Nakagawa S, Tezuka K, Akagi H, Kawamoto T. 2016. Breeding and characterization of the rice (Oryza sativa L.) line “Akita 110” for cadmium phytoremediation. Soil Sci Plant Nutr, 62(4): 373-378. |
| [122] | Tan J J, Wang J W, Chai T Y, Zhang Y X, Feng S S, Li Y, Zhao H J, Liu H M, Chai X P. 2013. Functional analyses of TaHMA2, a P1B-type ATPase in wheat. Plant Biotechnol J, 11(4): 420-431. |
| [123] | Tan L T, Zhu Y X, Fan T, Peng C, Wang J R, Sun L, Chen C Y. 2019. OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem Biophys Res Commun, 512(1): 112-118. |
| [124] |
Tan L T, Qu M M, Zhu Y X, Peng C, Wang J R, Gao D Y, Chen C Y. 2020. ZINC TRANSPORTER5 and ZINC TRANSPORTER9 function synergistically in zinc/cadmium uptake. Plant Physiol, 183(3): 1235-1249.
DOI PMID |
| [125] | Tanaka N, Uraguchi S, Kajikawa M, Saito A, Ohmori Y, Fujiwara T. 2018. A rice PHD-finger protein OsTITANIA, is a growth regulator that functions through elevating expression of transporter genes for multiple metals. Plant J, 96(5): 997-1006. |
| [126] |
Tang B, Luo M J, Zhang Y X, Guo H L, Li J N, Song W, Zhang R Y, Feng Z, Kong M S, Li H, Cao Z Y, Lu X D, Li D L, Zhang J H, Wang R H, Wang Y D, Chen Z H, Zhao Y X, Zhao J R. 2021. Natural variations in the P-type ATPase heavy metal transporter gene ZmHMA3 control cadmium accumulation in maize grains. J Exp Bot, 72(18): 6230-6246.
DOI PMID |
| [127] | Tang L, Mao B G, Li Y K, Lv Q M, Zhang L P, Chen C Y, He H J, Wang W P, Zeng X F, Shao Y, Pan Y L, Hu Y Y, Peng Y, Fu X Q, Li H Q, Xia S T, Zhao B R. 2017. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci Rep, 7(1): 14438. |
| [128] |
Tang W, Charles T M, Newton R J. 2005. Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus virginiana mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol Biol, 59(4): 603-617.
DOI PMID |
| [129] |
Thomine S, Wang R, Ward J M, Crawford N M, Schroeder J I. 2000. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA, 97(9): 4991-4996.
DOI PMID |
| [130] |
Thomine S, Lelièvre F, Debarbieux E, Schroeder J I, Barbier- Brygoo H. 2003. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J, 34(5): 685-695.
DOI PMID |
| [131] | Tian S Q, Liang S, Qiao K, Wang F H, Zhang Y X, Chai T Y. 2019. Co-expression of multiple heavy metal transporters changes the translocation, accumulation, and potential oxidative stress of Cd and Zn in rice (Oryza sativa). J Hazard Mater, 380: 120853. |
| [132] | Tran T A, Popova L P. 2013. Functions and toxicity of cadmium in plants: Recent advances and future prospects. Turk J Bot, 37(1): 1-13. |
| [133] |
Ueno D, Kono I, Yokosho K, Ando T, Yano M, Ma J F. 2009. A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa). New Phytol, 182(3): 644-653.
DOI PMID |
| [134] |
Ueno D, Yamaji N, Kono I, Huang C F, Ando T, Yano M, Ma J F. 2010. Gene limiting cadmium accumulation in rice. Proc Natl Acad Sci USA, 107(38): 16500-16505.
DOI PMID |
| [135] |
Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S. 2009. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J Exp Bot, 60(9): 2677-2688.
DOI PMID |
| [136] |
Uraguchi S, Kamiya T, Sakamoto T, Kasai K, Sato Y, Nagamura Y, Yoshida A, Kyozuka J, Ishikawa S, Fujiwara T. 2011. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc Natl Acad Sci USA, 108(52): 20959-20964.
DOI PMID |
| [137] |
Verbruggen N, Hermans C, Schat H. 2009. Mechanisms to cope with arsenic or cadmium excess in plants. Curr Opin Plant Biol, 12(3): 364-372.
DOI PMID |
| [138] | Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot M L, Briat J F, Curie C. 2002. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell, 14(6): 1223-1233. |
| [139] | Vijayaragavan M, Prabhahar C, Sureshkumar J, Natarajan A, Vijayarengan P, Sharavanan S. 2011. Toxic effect of cadmium on seed germination, growth and biochemical contents of cowpea (Vigna unguiculata l.) plants. Int Multidiscip Res J, 1/5: 1-6. |
| [140] | Vinocur B, Altman A. 2005. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr Opin Biotechnol, 16(2): 123-132. |
| [141] | Visarada K B R S, Meena K, Aruna C, Srujana S, Saikishore N, Seetharama N. 2009. Transgenic breeding: Perspectives and prospects. Crop Sci, 49(5): 1555-1563. |
| [142] | Wang T K, Li Y X, Fu Y F, Xie H J, Song S F, Qiu M D, Wen J, Chen M W, Chen G, Tian Y, Li C X, Yuan D Y, Wang J L, Li L. 2019. Mutation at different sites of metal transporter gene OsNramp5 affects Cd accumulation and related agronomic traits in rice (Oryza sativa L.). Front Plant Sci, 10: 1081. |
| [143] | Wang X X, Gao Y, Feng Y, Li X, Wei Q, Sheng X Y. 2014. Cadmium stress disrupts the endomembrane organelles and endocytosis during Picea wilsonii pollen germination and tube growth. PLoS One, 9(4): e94721. |
| [144] | Wang Y, Qian Y R, Hu H, Xu Y, Zhang H J. 2011. Comparative proteomic analysis of Cd-responsive proteins in wheat roots. Acta Physiol Plant, 33(2): 349-357. |
| [145] |
Wang Y, Yu K F, Poysa V, Shi C, Zhou Y H. 2012. A single point mutation in GmHMA3 affects cadmium (Cd) translocation and accumulation in soybean seeds. Mol Plant, 5(5): 1154-1156.
DOI PMID |
| [146] |
Wang Y, Wang C, Liu Y J, Yu K F, Zhou Y H. 2018. GmHMA3 sequesters Cd to the root endoplasmic reticulum to limit translocation to the stems in soybean. Plant Sci, 270: 23-29.
DOI PMID |
| [147] | White P J, Brown P H. 2010. Plant nutrition for sustainable development and global health. Ann Bot, 105(7): 1073-1080. |
| [148] |
Wu D Z, Yamaji N, Yamane M, Kashino-Fujii M, Sato K, Ma J F. 2016. The HvNramp5 transporter mediates uptake of cadmium and manganese, but not iron. Plant Physiol, 172(3): 1899-1910.
PMID |
| [149] | Wu H L, Chen C L, Du J, Liu H F, Cui Y, Zhang Y, He Y J, Wang Y Q, Chu C C, Feng Z Y, Li J M, Ling H Q. 2012. Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol, 158(2): 790-800. |
| [150] | Wu S W, Hu C X, Wang X M, Wang Y W, Yu M, Xiao H D, Shabala S, Wu K J, Tan Q L, Xu S J, Sun X C. 2022. Cadmium- induced changes in composition and co-metabolism of glycerolipids species in wheat root: Glycerolipidomic and transcriptomic approach. J Hazard Mater, 423: 127115. |
| [151] | Xie H L, Jiang R F, Zhang F S, McGrath S P, Zhao F J. 2009. Effect of nitrogen form on the rhizosphere dynamics and uptake of cadmium and zinc by the hyperaccumulator Thlaspi caerulescens. Plant Soil, 318: 205-215. |
| [152] | Xiong Z T, Peng Y H. 2001. Response of pollen germination and tube growth to cadmium with special reference to low concentration exposure. Ecotoxicol Environ Saf, 48(1): 51-55. |
| [153] | Xu W H, Li W Y, He J P, Balwant S, Xiong Z T. 2009. Effects of insoluble Zn, Cd, and EDTA on the growth, activities of antioxidant enzymes and uptake of Zn and Cd in Vetiveria zizanioides. J Environ Sci, 21(2): 186-192. |
| [154] |
Yamaji N, Xia J X, Mitani-Ueno N, Yokosho K, Ma J F. 2013. Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2. Plant Physiol, 162(2): 927-939.
DOI PMID |
| [155] | Yan H L, Xu W X, Xie J Y, Gao Y W, Wu L L, Sun L, Chen X, Zhang T, Dai C H, Lin X N, Feng L, Wang X Q, Li F M, Zhu X Y, Li J J, Li Z C, Chen C Y, Ma M, Zhang H L, He Z Y. 2018. Natural variation OsCd1V449 contributes to reducing cadmium accumulation in rice grain. Preprints, 2018: 2018020075. |
| [156] | Yan H L, Xu W X, Xie J Y, Gao Y W, Wu L L, Sun L, Feng L, Chen X, Zhang T, Dai C H, Li T, Lin X N, Zhang Z Y, Wang X Q, Li F M, Zhu X Y, Li J J, Li Z C, Chen C Y, Ma M, Zhang H L, He Z Y. 2019. Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies. Nat Commun, 10(1): 2562. |
| [157] | Yang C H, Zhang Y, Huang C F. 2019. Reduction in cadmium accumulation in japonica rice grains by CRISPR/Cas9-mediated editing of OsNRAMP5. J Integr Agric, 18(3): 688-697. |
| [158] | Yang Y J, Xiong J, Tao L X, Cao Z Z, Tang W, Zhang J P, Yu X Y, Fu G F, Zhang X F, Lu Y L. 2020. Regulatory mechanisms of nitrogen (N) on cadmium (Cd) uptake and accumulation in plants: A review. Sci Total Environ, 708: 135186. |
| [159] | Younis U, Malik S A, Rizwan M, Qayyum M F, Ok Y S, Shah M H R, Rehman R A, Ahmad N. 2016. Biochar enhances the cadmium tolerance in spinach (Spinacia oleracea) through modification of Cd uptake and physiological and biochemical attributes. Environ Sci Pollut Res Int, 23(21): 21385-21394. |
| [160] | Yu H, Wang J L, Fang W, Yuan J G, Yang Z Y. 2006. Cadmium accumulation in different rice cultivars and screening for pollution- safe cultivars of rice. Sci Total Environ, 370(2/3): 302-309. |
| [161] | Zeng T, Fang B H, Huang F L, Dai L, Tang Z, Tian J L, Cao G D, Meng X L, Liu Y C, Lei B, Lu M H, Cai Z W. 2021. Mass spectrometry-based metabolomics investigation on two different indica rice grains (Oryza sativa L.) under cadmium stress. Food Chem, 343: 128472. |
| [162] | Zhang D Z, Zhou H, Shao L L, Wang H R, Zhang Y B, Zhu T, Ma L T, Ding Q, Ma L J. 2022. Root characteristics critical for cadmium tolerance and reduced accumulation in wheat (Triticum aestivum L.). J Environ Manage, 305: 114365. |
| [163] | Zhang F G, Liu M H, Li Y, Che Y Y, Xiao Y. 2019. Effects of arbuscular mycorrhizal fungi, biochar and cadmium on the yield and element uptake of Medicago sativa. Sci Total Environ, 655: 1150-1158. |
| [164] | Zhang J L, Zhu Y C, Yu L J, Yang M, Zou X, Yin C X, Lin Y J. 2022. Research advances in cadmium uptake, transport and resistance in rice (Oryza sativa L.). Cells, 11(3): 569. |
| [165] | Zhang L Y, Zhang H Y, Guo W, Tian Y L, Chen Z S, Wei X F. 2012. Photosynthetic responses of energy plant maize under cadmium contamination stress. Adv Mater Res, 356/360: 283-286. |
| [166] |
Zhang W X, Guan M Y, Wang J, Wang Y L, Zhang W G, Lu X Z, Xu P, Chen M X, Zhu Y W. 2023. Screening rice germplasm with different genetic backgrounds for cadmium accumulation in brown rice in cadmium-polluted soils. Rice Sci, 30(4): 267-270.
DOI |
| [167] | Zhao F J, Huang X Y. 2018. Cadmium phytoremediation: Call rice CAL1. Mol Plant, 11(5): 640-642. |
| [168] |
Zhu X F, Zhao L, Huang J, He J T, Song J Y, Teng Y, Shen R F. 2024. Cell wall fixation, translocation, and vacuolar detoxification of cadmium contribute to differential grain cadmium accumulation in two rice cultivars. Rice Sci, 31(3): 241-244.
DOI |
| [169] | Zulfiqar U, Farooq M, Hussain S, Maqsood M, Hussain M, Ishfaq M, Ahmad M, Anjum M Z. 2019. Lead toxicity in plants: Impacts and remediation. J Environ Manage, 250: 109557. |
| [1] | Zhou Tianshun, Yu Dong, Wu Liubing, Xu Yusheng, Duan Meijuan, Yuan Dingyang. Seed Storability in Rice: Physiological Foundations, Molecular Mechanisms, and Applications in Breeding [J]. Rice Science, 2024, 31(4): 401-416. |
| [2] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [3] | Ning Xiao, Yunyu Wu, Aihong Li. Strategy for Use of Rice Blast Resistance Genes in Rice Molecular Breeding [J]. Rice Science, 2020, 27(4): 263-277. |
| [4] | ZHANG Chun-hua1, 2, WU Ze-ying3, JU Ting3, GE Ying2, 3. Purification and Identification of Glutathione S-transferase in Rice Root under Cadmium Stress [J]. RICE SCIENCE, 2013, 20(3): 173-178. |
| [5] | ZHANG Chun-hua , GE Ying . Response of Glutathione and Glutathione S-transferase in Rice Seedlings Exposed to Cadmium Stress [J]. RICE SCIENCE, 2008, 15(1): 73-76 . |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||