
Rice Science ›› 2018, Vol. 25 ›› Issue (4): 227-234.DOI: 10.1016/j.rsci.2018.06.006
• Orginal Article • Previous Articles
Kaizhuan Xiao1,2,3, Xiaohui Mao2,3, Yingheng Wang2,3, Jinlan Wang2,3, Yidong Wei2,3, Qiuhua Cai2,3, Hua’an Xie1,2,3( ), Jianfu Zhang1,2,3(
), Jianfu Zhang1,2,3( )
)
Received:2017-12-21
Accepted:2018-04-20
Online:2018-06-20
Published:2018-04-10
Kaizhuan Xiao, Xiaohui Mao, Yingheng Wang, Jinlan Wang, Yidong Wei, Qiuhua Cai, Hua’an Xie, Jianfu Zhang. Transcript Profiling Reveals Abscisic Acid, Salicylic Acid and Jasmonic-Isoleucine Pathways Involved in High Regenerative Capacities of Immature Embryos Compared with Mature Seeds in japonica Rice[J]. Rice Science, 2018, 25(4): 227-234.
Add to citation manager EndNote|Ris|BibTeX
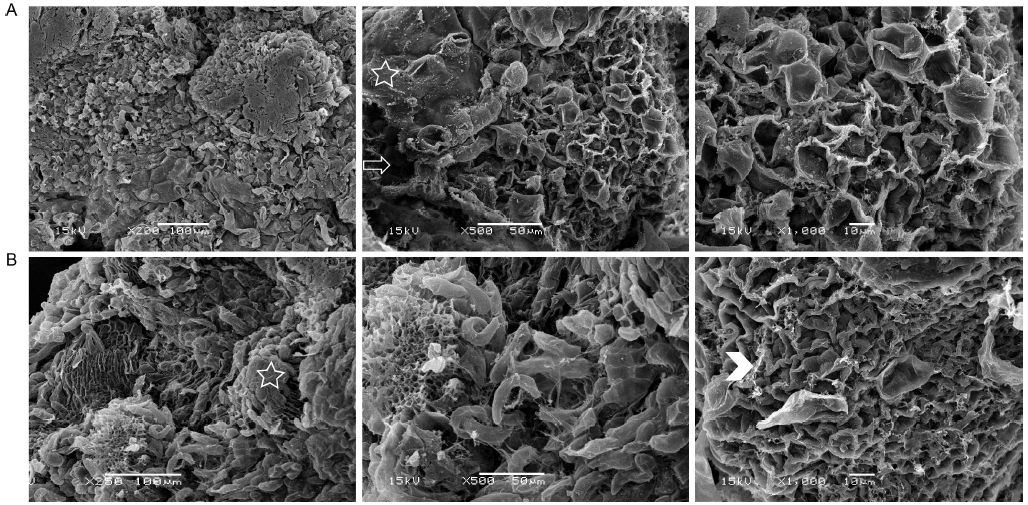
Fig. 1. SEM images of callus surface of IMN (A) and MN (B) after the placement of the callus induction medium for 20 d. SEM, Scanning electron microscopy; IMN, Immature Nipponbare embryos; MN, Mature Nipponbare seeds.The asterisks and arrowheads indicate the membrane and fibrillar of the extracellular matrix of the callus respectively. The arrow indicates the membrane structures with holes covering the callus surface.
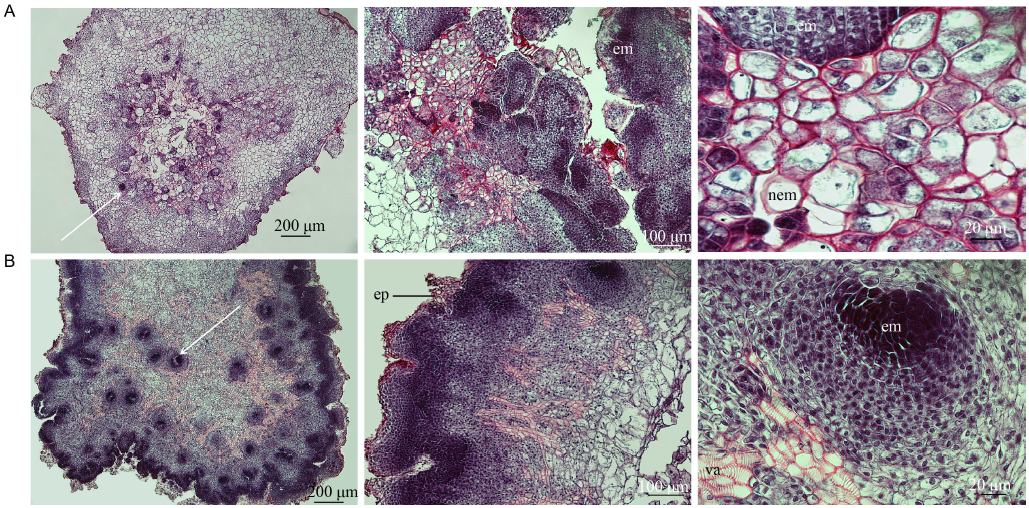
Fig. 2. Histochemical analysis of the somatic embryogenesis of IMN (A) and MN (B) after placement on callus induction medium for 20 d. IMN, Immature Nipponbare embryos; MN, Mature Nipponbare seeds. The em, nem and white arrows indicate the embryogenic callus units, nonembryogenic callus and large callus domain/protuberances. The va and ep represent the vascellum and epidermis-like tissue on the surface of some protuberances, respectively.
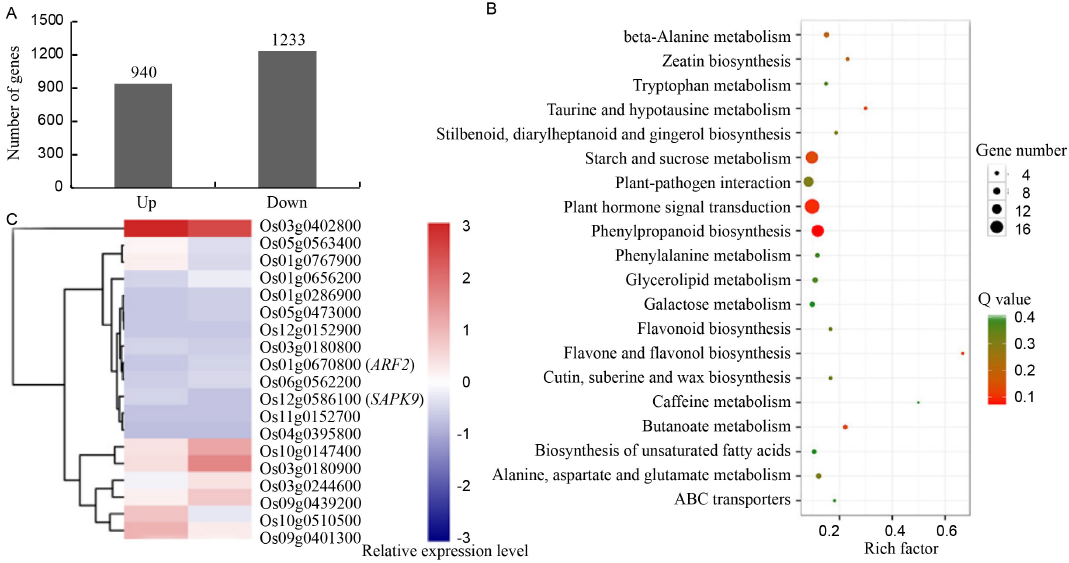
Fig. 3. Global analyze of the mRNA transcriptome data. A, Differentially expressed genes (DEGs) between immature embryos and mature seeds involved in callus induction. B, Kyoto encyclopedia of genes and genomes (KEGG) pathway abundance of DEGs. A total of 20 pathways were identified in each comparison based on 0 < Q value < 1.0, where the abundance of the pathway is more significant if the value is close to zero. Y-axis shows the pathway categories, and X-axis shows the richness factor of each pathway. C, The heat map of genes involved in plant hormone signal transduction pathway.
| Sample ID | Total read | GC content (%) | Data with Phred score ≥ 20 (%) | Data with Phred score ≥ 30 (%) | Number of reads aligned to reference genome (%) |
|---|---|---|---|---|---|
| MN-1 | 29 477 712 | 51.96 | 96.37 | 91.27 | 26 431 504 (89.67) |
| MN-2 | 30 345 732 | 51.95 | 96.04 | 90.60 | 26 884 157 (88.59) |
| MN-3 | 31 118 488 | 52.04 | 96.08 | 90.67 | 27 392 769 (88.03) |
| IMN-1 | 33 653 030 | 51.82 | 96.29 | 91.23 | 30 135 270 (89.55) |
| IMN-2 | 37 035 338 | 53.44 | 96.03 | 90.87 | 32 262 197 (87.11) |
| IMN-3 | 29 646 958 | 51.47 | 96.14 | 91.00 | 26 431 890 (89.16) |
| IMN, Immature Nipponbare embryos; MN, Mature Nipponbare seeds. | |||||
Table 1 Summary of the fastq file generated by individually sequencing the RNA of all samples.
| Sample ID | Total read | GC content (%) | Data with Phred score ≥ 20 (%) | Data with Phred score ≥ 30 (%) | Number of reads aligned to reference genome (%) |
|---|---|---|---|---|---|
| MN-1 | 29 477 712 | 51.96 | 96.37 | 91.27 | 26 431 504 (89.67) |
| MN-2 | 30 345 732 | 51.95 | 96.04 | 90.60 | 26 884 157 (88.59) |
| MN-3 | 31 118 488 | 52.04 | 96.08 | 90.67 | 27 392 769 (88.03) |
| IMN-1 | 33 653 030 | 51.82 | 96.29 | 91.23 | 30 135 270 (89.55) |
| IMN-2 | 37 035 338 | 53.44 | 96.03 | 90.87 | 32 262 197 (87.11) |
| IMN-3 | 29 646 958 | 51.47 | 96.14 | 91.00 | 26 431 890 (89.16) |
| IMN, Immature Nipponbare embryos; MN, Mature Nipponbare seeds. | |||||
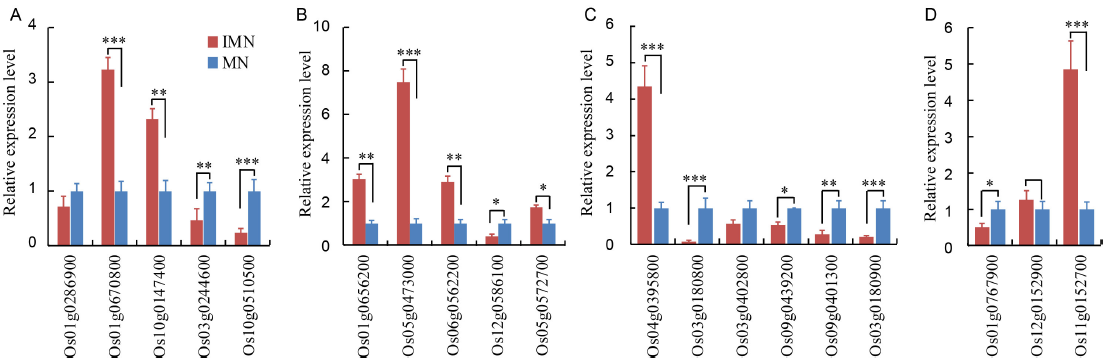
Fig. 4. Relative expression profiles of genes involved in plant hormone signal transduction pathway validated with quantitative real time PCR with the ΔΔCT method. A, Auxin relative genes; B, Abscisic acid relative genes; C, Jasmonic acid relative genes; D, Salicylic acid relative genes.IMN, Immature Nipponbare embryos; MN, Mature Nipponbare seeds.Data represent mean ± SE (n = 5). The significant differences between mean values are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001) using the Student’s t-test.
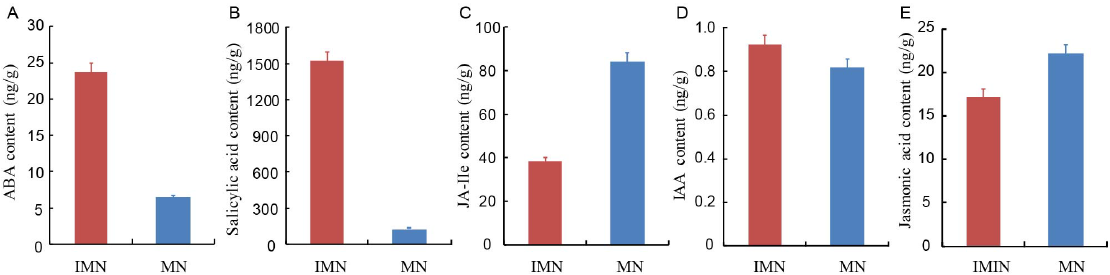
Fig. 5. Abscisic acid (ABA, A), salicylic acid (B), jasmonoyl-isoleucine (JA-Ile, C), indole-3-acetic acid (IAA, D), and jasmonic acid (E) contents of immature Nippobare embryos (IMN) and mature Nippobare seeds (MN).Data represent mean ± SE (n = 5).
| [1] | Ahmadi B, Shariatpanahi M E, da Silva J A T.2014. Efficient induction of microspore embryogenesis using abscisic acid, jasmonic acid and salicylic acid inBrassica napus L. Plant Cell Tiss Org Cult, 116(3): 343-351. |
| [2] | Chu Z L, Chen J Y, Xu H X, Dong Z D, Chen F, Cui D Q.2016. Identification and comparative analysis of microRNA in wheat (Triticum aestivum L.) callus derived from mature and immature embryos during in vitro culture. Front Plant Sci, 7(1119): 1302. |
| [3] | da Silva J A T.2013. Jasmonic acid, but not salicylic acid, improves PLB formation of hybrid Cymbidium.Plant Cell Tiss Org Cult, 22: 187-192. |
| [4] | Duan Y B, Zhai C G, Li H, Li H, Li J, Mei W Q, Gui H P, Ni D H, Song F S, Li L, Zhang W G, Yang J B.2012. An efficient and high-throughput protocol forAgrobacterium-mediated transformation based on phosphomannose isomerase positive selection in japonica rice(Oryza sativa L.). Plant Cell Rep, 31: 1611-1624. |
| [5] | Hiei Y, Ohta S, Komari T, Kumashiro T.1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J, 6(2): 271-282. |
| [6] | Hiei Y, Komari T.2008. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed.Nat Protoc, 3: 824-834. |
| [7] | Hofmann N R.2016. A breakthrough in monocot transformation methods.Plant Cell, 28: 1989. |
| [8] | Hoque M E, Mansfield J W, Bennett M H.2005. Agrobacterium- mediated transformation ofindica rice genotypes: An assessment of factors affecting the transformation efficiency. Plant Cell Tiss Org Cult, 82(1): 45-55. |
| [9] | Hu L, Li H, Qin R Y, Xu R F, Li J, Li L, Wei P C, Yang J B.2016. Plant phosphomannose isomerase as a selectable marker for rice transformation.Sci Rep, 6: 25921. |
| [10] | Islam M M, Roly Z Y, Lee Y, Khalekuzzaman M.2014. In vitro propagation and genetic transformation system using immature embryo in elite rice (Oryza sativa L.) cultivars. Plant Breed Biotechnol, 2(1): 88-96. |
| [11] | Jena K K, Hechanova S L, Verdeprado H, Prahalada G D, Kim S R.2017. Development of 25 near-isogenic lines (NILs) with ten BPH resistance genes in rice (Oryza sativa L.): Production, resistance spectrum, and molecular analysis. Theor Appl Genet, 130: 1-16. |
| [12] | Jimenez V M, Thomas C.2006. Participation of plant hormones in determination and progression of somatic embryogenesis.Plant Cell Monogr, 2: 103-118. |
| [13] | Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S L.2013. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions.Genome Biol, 14(4): 36. |
| [14] | Kim J, Harter K, Theologis A.1997. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA, 94: 11786-11791. |
| [15] | Kumar V, Moyo M, van Staden J.2015. Somatic embryogenesis ofPelargonium sidoides DC. Plant Cell Tiss Org Cult, 121: 571-577. |
| [16] | Liu J F, Cheng Y Q, Yan K, Liu Q, Wang Z W.2012. The relationship between reproductive growth and blank fruit formation inCorylus heterophylla Fisch. Sci Hort, 136: 128-134. |
| [17] | Mao X Z, Cai T, Olyarchuk J G, Wei L P.2005. Automated genome annotation and pathway identification using KEGG orthology (KO) as a controlled vocabulary.Bioinformatics, 21: 3787-3793. |
| [18] | Mullet J E.2017. High-biomass C4 grasses-filling the yield gap.Plant Sci, 261: 10-17. |
| [19] | Perez M, Canal M J, Toorop P E.2015. Expression analysis of epigenetic and abscisic acid-related genes during maturation ofQuercus suber somatic embryos. Plant Cell Tiss Org Cult, 121(2): 353-366. |
| [20] | Rikiishi K, Matsuura T, Ikeda Y, Maekawa M.2015. Light inhibition of shoot regeneration is regulated by endogenous abscisic acid level in calli derived from immature barley embryos.PLoS One, 10: e0145242. |
| [21] | Rudus I, Kępczynski J, Kępczynska E.2001. The influence of the jasmonates and abscisic acid on callus growth and somatic embryogenesis inMedicago sativa L. tissue culture. Acta Physiol Plant, 23: 103-107. |
| [22] | Shen L, Li J, Fu Y P, Wang J J, Hua Y F, Jiao X Z, Yan C J, Wang K J.2017. Orientation improvement of grain length and grain number in rice by using CRISPR/Cas9 system.Chin J Rice Sci, 31(3): 223-231. (in Chinese with English abstract) |
| [23] | Slamet-Loedin I H, Chadha-Mohanty P, Torrizo L.2014. Agrobacterium-mediated transformation: Rice transformation.Cereal Genom, 1099: 261-271. |
| [24] | Stitz M, Gase K, Baldwin I T, Gaquerel E.2011. Ectopic expression ofAtJMT in Nicotiana attenuata: Creating a metabolic sink has tissue-specific consequences for the jasmonate metabolic network and silences downstream gene expression. J Plant Physiol, 157(1): 341-354. |
| [25] | Tiwari S B, Hagen G, Guilfoyle T J.2004. Aux/IAA proteins contain a potent transcriptional repression domain.Plant Cell, 16(2): 533-543. |
| [26] | Toriyama K, Arimoto Y, Uchimiya H, Hinata K.1988. Transgenic rice plants after direct gene transfer into protoplasts.Nat Biotechnol, 6: 1072-1074. |
| [27] | Trapnell C, Williams B A, Pertea G, Mortazavi A, Kwan G, van Baren M J, Salzberg S L, Wold B J, Pachter L.2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation.Nat Biotechnol, 28(5): 511-515. |
| [28] | Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley D R, Pimentel H, Salzberg S L, Rinn J L, Pachter L.2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks.Nat Protoc, 7(3): 562-578. |
| [29] | Visser C, Qureshi J A, Gill R, Saxena P X.1992. Morpho- regulatory role of thidiazuron substitution of auxin and cytokinin requirement for the induction of somatic embryogenesis in geranium hypocotyl cultures.J Plant Physiol, 99(4): 1704-1707. |
| [30] | Wang L, Feng Z, Wang X, Wang X, Zhang X.2010. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data.Bioinformatics, 26(1): 136-138. |
| [31] | Yang H, Zhang H M, Davey M R, Mulligan B J, Cocking E C.1988. Production of kanamycin resistant rice tissues following DNA uptake into protoplasts.Plant Cell Rep, 7(6): 421-425. |
| [32] | Yang X Q, Chen T T, Zhao X, Zhang C X, Yang Y J, Fu G F, Tao L X.2016. Mechanism behind the effects of GA3 and PP33 on grain yield formation of super rice Yongyou 12.Chin J Rice Sci, 30(1): 53-61. (in Chinese with English abstract) |
| [33] | Yang X Y, Zhang X L.2010. Regulation of somatic embryogenesis in higher plants.Crit Rev Plant Sci, 29: 36-57. |
| [34] | Yang X Y, Zhang X L, Yuan D J, Jin F Y, Zhang Y C, Xu J.2012. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton.BMC Plant Biol, 12: 110. |
| [35] | Zhang G Y, Liu R R, Xu G, Zhang P, Li Y, Tang K X, Liang G H, Liu Q Q.2013. Increased α-tocotrienol content in seeds of transgenic rice over-expressingArabidopsis γ-tocopherol methyltransferase. Transgenic Res, 22(1): 89-99. |
| [36] | Zhang H M, Yang H, Rech E L, Golds T J, Davis A S, Mulligan B J, Cocking E C, Davey M R.1988. Transgenic rice plants produced by electroporation-mediated plasmid uptake into protoplasts.Plant Cell Rep, 7(6): 379-384. |
| [37] | Zhang L H, Chen H W, Li Y L, Wang S J, Su J P, Liu X J, Chen D F, Chen X W.2014. Evaluation of the agronomic performance of atrazine-tolerant transgenicjaponica rice parental lines for utilization in hybrid seed production. PLoS One, 9: e108569. |
| [1] | Jiang Changjie, Liang Zhengwei, Xie Xianzhi. Priming for Saline-Alkaline Tolerance in Rice: Current Knowledge and Future Challenges [J]. Rice Science, 2023, 30(5): 417-425. |
| [2] | Chen Yanhua, Wang Yaliang, Chen Huizhe, Xiang Jing, Zhang Yikai, Wang Zhigang, Zhu Defeng, Zhang Yuping. Brassinosteroids Mediate Endogenous Phytohormone Metabolism to Alleviate High Temperature Injury at Panicle Initiation Stage in Rice [J]. Rice Science, 2023, 30(1): 70-86. |
| [3] | M. Iqbal R. Khan, Sarika Kumari, Faroza Nazir, Risheek Rahul Khanna, Ravi Gupta, Himanshu Chhillar. Defensive Role of Plant Hormones in Advancing Abiotic Stress-Resistant Rice Plants [J]. Rice Science, 2023, 30(1): 15-35. |
| [4] | Ersong Zheng, Xuming Wang, Rumeng Xu, Feibo Yu, Chao Zheng, Yong Yang, Yang Chen, Jianping Chen, Chengqi Yan, Jie Zhou. Regulation of OsPR10a Promoter Activity by Phytohormone and Pathogen Stimulation in Rice [J]. Rice Science, 2021, 28(5): 442-456. |
| [5] | Ramakrishna Wusirika, Kumari Anuradha, Rahman Nafeesa, Mandave Pallavi. Anticancer Activities of Plant Secondary Metabolites: Rice Callus Suspension Culture as a New Paradigm [J]. Rice Science, 2021, 28(1): 13-30. |
| [6] | Eo Seong-Hui, Ja Kim Song. Iksan526 Rice Callus Extract Induces Dedifferentiation of Rabbit Articular Chondrocytes via ERK1/2 and PI-3K/Akt Pathways [J]. Rice Science, 2020, 27(6): 504-514. |
| [7] | Yongqi He, Jia Zhao, Defeng Feng, Zhibo Huang, Jiaming Liang, Yufei Zheng, Jinping Cheng, Jifeng Ying, Zhoufei Wang. RNA-Seq Study Reveals AP2-Domain-Containing Signalling Regulators Involved in Initial Imbibition of Seed Germination in Rice [J]. Rice Science, 2020, 27(4): 302-314. |
| [8] | Yanjie Xu, Yining Ying, Shuhong Ouyang, Xiaoliang Duan, Hui Sun, Shukun Jiang, Shichen Sun, Jinsong Bao. Factors Affecting Sensory Quality of Cooked japonica Rice [J]. Rice Science, 2018, 25(6): 330-339. |
| [9] | Radhesh Krishnan Subramanian, Muthuramalingam Pandiyan, Pandian Subramani, Banupriya Ramachandradoss, Chithra Gunasekar, Ramesh Manikandan. Sprouted Sorghum Extract Elicits Coleoptile Emergence, Enhances Shoot and Root Acclimatization, and Maintains Genetic Fidelity in indica Rice [J]. Rice Science, 2018, 25(2): 61-72. |
| [10] | Matusmoto Tadashi, Yamada Kazuhiro, Yoshizawa Yuko, Oh Keimei. Comparison of Effect of Brassinosteroid and Gibberellin Biosynthesis Inhibitors on Growth of Rice Seedlings [J]. Rice Science, 2016, 23(1): 51-55. |
| [11] | Pawar Bhausaheb, Kale Prashant, Bahurupe Jyoti, Jadhav Ashok, Kale Anil, Pawar Sharad. Proline and Glutamine Improve in vitro Callus Induction and Subsequent Shooting in Rice [J]. Rice Science, 2015, 22(6): 283-289. |
| [12] | Xiu-mei Wang, Yue-yang Liang, Ling Li, Chang-wei Gong, Hai-peng Wang, Xiao-xi Huang, Shuang-cheng Li, Qi-ming Deng, Jun Zhu, Ai-ping Zheng, Ping Li, Shi-quan Wang. Identification and Cloning of Tillering-Related Genes OsMAX1 in Rice [J]. Rice Science, 2015, 22(6): 255-263. |
| [13] | P. MANIMARAN, G. RAVI KUMAR, M. RAGHURAMI REDDY, S. JAIN, T. BHASKAR RAO, S. K. MANGRAUTHIA, R. M. SUNDARAM, S. RAVICHANDRAN, S. M. BALACHANDRAN. Infection of Early and Young Callus Tissues of Indica Rice BPT 5204 Enhances Regeneration and Transformation Efficiency [J]. RICE SCIENCE, 2013, 20(6): 415-426. |
| [14] | TIAN Fu-kuan1, 2, #, RUAN Ban-pu1, 3, #, YAN Mei-xian1, YE Shi-fang1, PENG You-lin1, DONG Guo-jun1, ZHU Li1, HU Jiang1, YAN Hong-lan1, GUO Long-biao1, QIAN Qian1, GAO Zhen-yu1. Genetic Analysis and QTL Mapping of Mature Seed Culturability in Indica Rice [J]. RICE SCIENCE, 2013, 20(5): 313-319. |
| [15] | FU Jing, XU Yun-ji, CHEN Lu, YUAN Li-min, WANG Zhi-qin, YANG Jian-chang. Changes in Enzyme Activities Involved in Starch Synthesis and Hormone Concentrations in Superior and Inferior Spikelets and Their Association with Grain Filling of Super Rice [J]. RICE SCIENCE, 2013, 20(2): 120-128. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||