
Rice Science ›› 2021, Vol. 28 ›› Issue (5): 427-430.DOI: 10.1016/j.rsci.2021.07.003
• Letter • Previous Articles Next Articles
Binte Monsur Mahmuda1,#, Ni Cao1,#, Xiangjin Wei1, Lihong Xie1, Guiai Jiao1, Shaoqing Tang1, Sreenivasulu Nese2, Gaoneng Shao1( ), Peisong Hu1(
), Peisong Hu1( )
)
Received:2020-10-11
Accepted:2021-03-04
Online:2021-09-28
Published:2021-09-28
About author:#These authors contributed equally to this work
Binte Monsur Mahmuda, Ni Cao, Xiangjin Wei, Lihong Xie, Guiai Jiao, Shaoqing Tang, Sreenivasulu Nese, Gaoneng Shao, Peisong Hu. Improved Eating and Cooking Quality of indica Rice Cultivar YK17 via Adenine Base Editing of Wxa Allele of Granule-Bound Starch Synthase I (GBSS I)[J]. Rice Science, 2021, 28(5): 427-430.
Add to citation manager EndNote|Ris|BibTeX
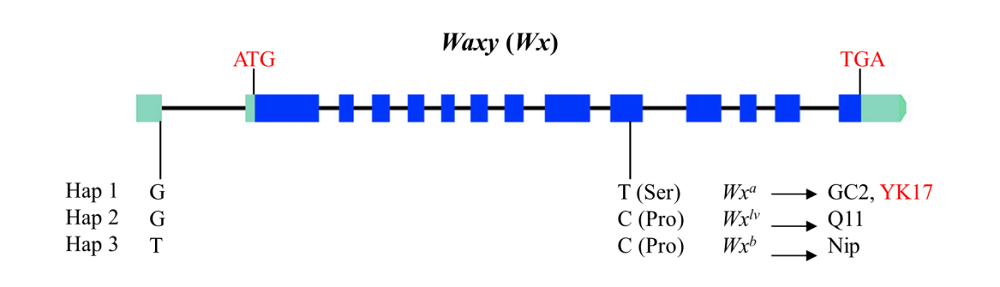
Fig. S1. Waxy genotype of YK17. Hap 1, Hap 2 and Hap 3 are three genotypes as described in Zhang et al (2019). GC2, Q11 and Nip are three rice accessions with different Waxy genotypes.
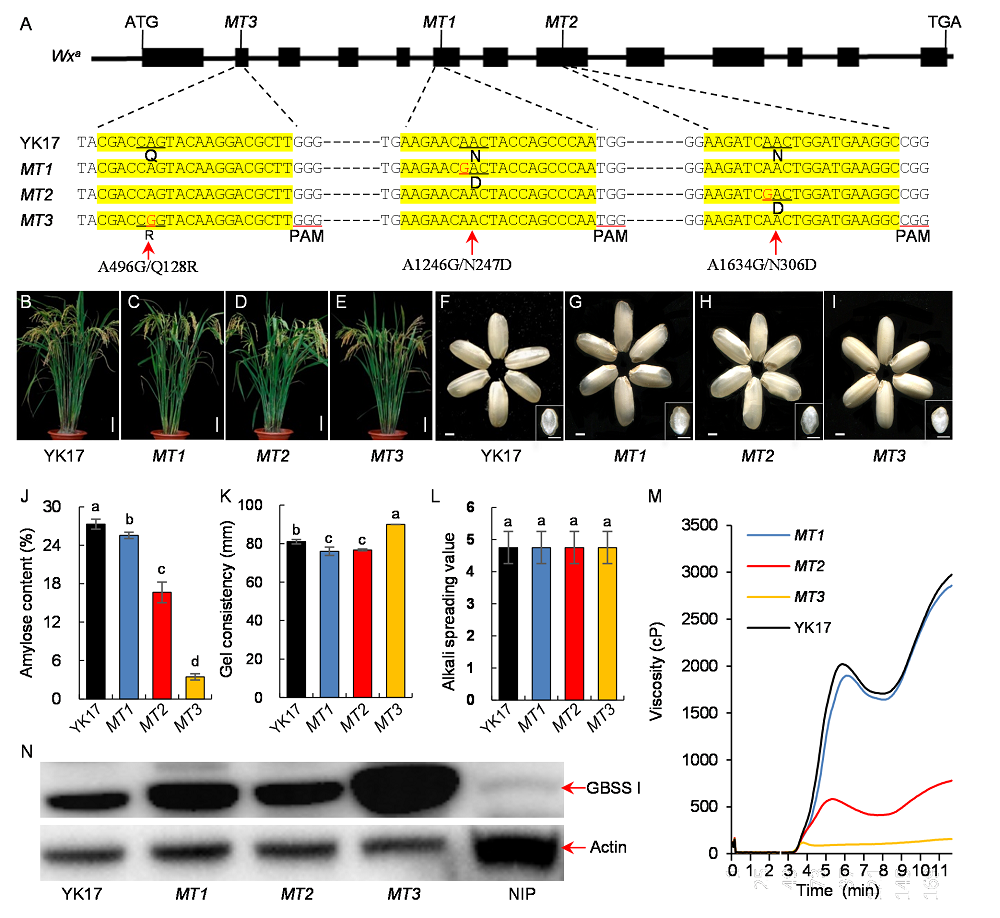
Fig. 1. Desirable amylose content of indica rice cultivar YK17 via adenine base editing of Wx gene. A, Structure of Wxa and the mutations in edited T1 lines. Protospacer-adjacent motifs (PAMs) include three bases (NGG) with a red underline. The red letters indicate altered bases. Q, Glutamine; R, Arginine; N, Asparagine; D, Aspartate. B-E, Gross morphologies of the wild type YK17 and its mutants MT1, MT2 and MT3. Scale bars are 10 cm. F-I, Appearance and transverse sections of brown rice of YK17 and mutants. Scale bars are 1 mm. J, Amylose content. K, Gel consistency. L, Alkali spreading values of the wild type and mutants. Data are Mean ± SD (n = 3) in J-L. Samples with different lowercase letters show a significant difference at P < 0.05 according to the Duncan’s test. M, Pasting properties of endosperm starch of YK17 and mutants. N, Western blotting of GBSS I in YK17 and mutant rice grains at 10 d after flowering. Actin was used as an internal control. NIP, Nipponbare; YK17, Zhongjiazao 17.
| Primer | Sequence (5' to 3') | For use |
|---|---|---|
| Waxy-target site 1-F | GGCGAAGAACAACTACCAGCCCAA | Vector construction |
| Waxy-target site 1-R | AAACTTGGGCTGGTAGTTGTTCTT | Vector construction |
| Waxy-target site 2-F | GGCGAAGATCAACTGGATGAAGGC | Vector construction |
| Waxy-target site 2-R | AAACGCCTTCATCCAGTTGATCTT | Vector construction |
| Waxy-target site 3-F | GGCGCGACCAGTACAAGGACGCTT | Vector construction |
| Waxy-target site 3-R | AAACAAGCGTCCTTGTACTGGTCG | Vector construction |
| Waxy-target site 1-seq-F | GGATGTTGTGTTCGTCTGCA | Target site sequencing |
| Waxy-target site 1-seq-R | AGGAGATGTTGTGGATGCAGA | Target site sequencing |
| Waxy-target site 2-seq-F | ACCCTGCACTACTGTCCATC | Target site sequencing |
| Waxy-target site 2-seq-R | CTAGGATCCCACTCGCTGAC | Target site sequencing |
| Waxy-target site 3-seq-F | TTGTGTTCTTTGCAGGCGAA | Target site sequencing |
| Waxy-target site 3-seq-R | TCGTACCTGTCTGCAACCTT | Target site sequencing |
| Target site 1-Chr1-seq-F | CGTTGCCCTACCATTGAAGA | Off-target sequencing |
| Target site 1-Chr1-seq-R | ACATTGGCTGCTGCTACTAC | Off-target sequencing |
| Target site 1-Chr5-seq-F | TGCGACAAGCTGTGGTATTT | Off-target sequencing |
| Target site 1-Chr5-seq-R | GCCTGTATGTAGCTCTGTTGA | Off-target sequencing |
| Target site 1-Chr11-seq-F | GTCATGAAGTGCCTGGAGAG | Off-target sequencing |
| Target site 1-Chr11-seq-R | TCGAAGAACATGAAGCGAGG | Off-target sequencing |
| Target site 1- Chr6-seq-F | GTAGCACCCAAAAGGAGCTT | Off-target sequencing |
| Target site 1- Chr6-seq-R | GTTTCTCCCCTTCTGCACTT | Off-target sequencing |
| Target site 1- Chr12-seq-F | TACTGGTGGTGAGGTTGTGA | Off-target sequencing |
| Target site 1- Chr12-seq-R | GGTAATATATGCATAGCAAACCTG | Off-target sequencing |
| Target site 2-Chr6-seq-F | CGCAAGTTACAGTTACAGGCT | Off-target sequencing |
| Target site 2-Chr6-seq-R | CTCCTTTCACCGGAAGATCC | Off-target sequencing |
| Target site 2-Chr5-seq-F | CCGATTAGGCGATTCTCCAG | Off-target sequencing |
| Target site 2-Chr5-seq-R | GCCAGGAAAACAGAAACAGC | Off-target sequencing |
| Target site 2-Chr3-seq-F | TGTCCAGCTAGATCCAACGA | Off-target sequencing |
| Target site 2-Chr3-seq-R | CGCCCTGATGTTGGTGTT | Off-target sequencing |
| Target site 2-Chr7-seq-F | TCAAGCACACACACAATCCA | Off-target sequencing |
| Target site 2-Chr7-seq-R | TCCAAATTTCAGCCACCGAA | Off-target sequencing |
| Target site 2-Chr9-seq-F | ACCATAAAGGTCACACCAGC | Off-target sequencing |
| Target site 2-Chr9-seq-R | CTCGGTTTAAACGTACTTACTGC | Off-target sequencing |
| Target site 3-Chr10-seq-F | CTCATCTTTCCTCCCCGTTG | Off-target sequencing |
| Target site 3-Chr10-seq-R | CTACTCCATCGCCTTATCAGC | Off-target sequencing |
| Target site 3-Chr2-seq-F | GCAATTGTGTGAACCATCGG | Off-target sequencing |
| Target site 3-Chr2-seq-R | ACCAGAAATTACTGCGCGAT | Off-target sequencing |
| Target site 3-Chr1-seq-F | GTCATCATCATGTACGGCGA | Off-target sequencing |
| Target site 3-Chr1-seq-R | TGCCGTTTATACTTGCGTGT | Off-target sequencing |
| Target site 3-Chr6-seq-F | TTTTACGTGCCCTTGAACCA | Off-target sequencing |
| Target site 3-Chr6-seq-R | ATCCATTAGAGGCAAGCGAG | Off-target sequencing |
| Target site 3-Chr9-seq-F | TGTCCTCTAGTTCTGGCGAA | Off-target sequencing |
| Target site 3-Chr9-seq-R | ATTTCTTGCCCCCACACAAT | Off-target sequencing |
Table S1 Primers used in this study.
| Primer | Sequence (5' to 3') | For use |
|---|---|---|
| Waxy-target site 1-F | GGCGAAGAACAACTACCAGCCCAA | Vector construction |
| Waxy-target site 1-R | AAACTTGGGCTGGTAGTTGTTCTT | Vector construction |
| Waxy-target site 2-F | GGCGAAGATCAACTGGATGAAGGC | Vector construction |
| Waxy-target site 2-R | AAACGCCTTCATCCAGTTGATCTT | Vector construction |
| Waxy-target site 3-F | GGCGCGACCAGTACAAGGACGCTT | Vector construction |
| Waxy-target site 3-R | AAACAAGCGTCCTTGTACTGGTCG | Vector construction |
| Waxy-target site 1-seq-F | GGATGTTGTGTTCGTCTGCA | Target site sequencing |
| Waxy-target site 1-seq-R | AGGAGATGTTGTGGATGCAGA | Target site sequencing |
| Waxy-target site 2-seq-F | ACCCTGCACTACTGTCCATC | Target site sequencing |
| Waxy-target site 2-seq-R | CTAGGATCCCACTCGCTGAC | Target site sequencing |
| Waxy-target site 3-seq-F | TTGTGTTCTTTGCAGGCGAA | Target site sequencing |
| Waxy-target site 3-seq-R | TCGTACCTGTCTGCAACCTT | Target site sequencing |
| Target site 1-Chr1-seq-F | CGTTGCCCTACCATTGAAGA | Off-target sequencing |
| Target site 1-Chr1-seq-R | ACATTGGCTGCTGCTACTAC | Off-target sequencing |
| Target site 1-Chr5-seq-F | TGCGACAAGCTGTGGTATTT | Off-target sequencing |
| Target site 1-Chr5-seq-R | GCCTGTATGTAGCTCTGTTGA | Off-target sequencing |
| Target site 1-Chr11-seq-F | GTCATGAAGTGCCTGGAGAG | Off-target sequencing |
| Target site 1-Chr11-seq-R | TCGAAGAACATGAAGCGAGG | Off-target sequencing |
| Target site 1- Chr6-seq-F | GTAGCACCCAAAAGGAGCTT | Off-target sequencing |
| Target site 1- Chr6-seq-R | GTTTCTCCCCTTCTGCACTT | Off-target sequencing |
| Target site 1- Chr12-seq-F | TACTGGTGGTGAGGTTGTGA | Off-target sequencing |
| Target site 1- Chr12-seq-R | GGTAATATATGCATAGCAAACCTG | Off-target sequencing |
| Target site 2-Chr6-seq-F | CGCAAGTTACAGTTACAGGCT | Off-target sequencing |
| Target site 2-Chr6-seq-R | CTCCTTTCACCGGAAGATCC | Off-target sequencing |
| Target site 2-Chr5-seq-F | CCGATTAGGCGATTCTCCAG | Off-target sequencing |
| Target site 2-Chr5-seq-R | GCCAGGAAAACAGAAACAGC | Off-target sequencing |
| Target site 2-Chr3-seq-F | TGTCCAGCTAGATCCAACGA | Off-target sequencing |
| Target site 2-Chr3-seq-R | CGCCCTGATGTTGGTGTT | Off-target sequencing |
| Target site 2-Chr7-seq-F | TCAAGCACACACACAATCCA | Off-target sequencing |
| Target site 2-Chr7-seq-R | TCCAAATTTCAGCCACCGAA | Off-target sequencing |
| Target site 2-Chr9-seq-F | ACCATAAAGGTCACACCAGC | Off-target sequencing |
| Target site 2-Chr9-seq-R | CTCGGTTTAAACGTACTTACTGC | Off-target sequencing |
| Target site 3-Chr10-seq-F | CTCATCTTTCCTCCCCGTTG | Off-target sequencing |
| Target site 3-Chr10-seq-R | CTACTCCATCGCCTTATCAGC | Off-target sequencing |
| Target site 3-Chr2-seq-F | GCAATTGTGTGAACCATCGG | Off-target sequencing |
| Target site 3-Chr2-seq-R | ACCAGAAATTACTGCGCGAT | Off-target sequencing |
| Target site 3-Chr1-seq-F | GTCATCATCATGTACGGCGA | Off-target sequencing |
| Target site 3-Chr1-seq-R | TGCCGTTTATACTTGCGTGT | Off-target sequencing |
| Target site 3-Chr6-seq-F | TTTTACGTGCCCTTGAACCA | Off-target sequencing |
| Target site 3-Chr6-seq-R | ATCCATTAGAGGCAAGCGAG | Off-target sequencing |
| Target site 3-Chr9-seq-F | TGTCCTCTAGTTCTGGCGAA | Off-target sequencing |
| Target site 3-Chr9-seq-R | ATTTCTTGCCCCCACACAAT | Off-target sequencing |
| Target | Homozygous | Heterozygous | Wild type | Total plants | Mutation efficiency (%) |
|---|---|---|---|---|---|
| Target site 1 (MT1) | 2 | 16 | 8 | 26 | 69.23 |
| Target site 2 (MT2) | 1 | 8 | 15 | 24 | 37.50 |
| Target site 3 (MT3) | 3 | 10 | 16 | 29 | 44.83 |
Table S2 Mutation efficiency in T0 generation.
| Target | Homozygous | Heterozygous | Wild type | Total plants | Mutation efficiency (%) |
|---|---|---|---|---|---|
| Target site 1 (MT1) | 2 | 16 | 8 | 26 | 69.23 |
| Target site 2 (MT2) | 1 | 8 | 15 | 24 | 37.50 |
| Target site 3 (MT3) | 3 | 10 | 16 | 29 | 44.83 |
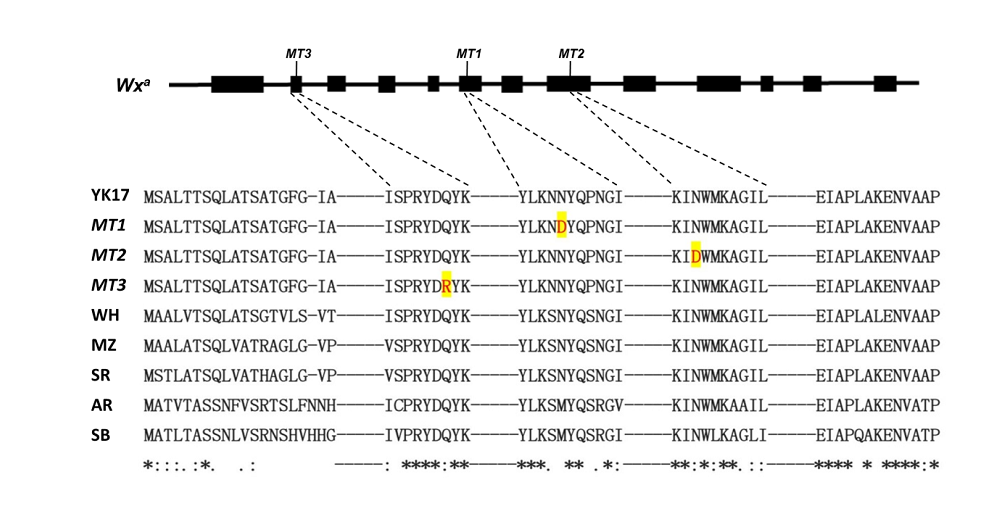
Fig. S4. Alignment of amino acids of Wxa mutants MT1, MT2 and MT3 conserved in different plant species.MT1 to MT3, Three mutants; WH, Wheat; MZ, Maize; SR, Sorghum; AR, Arabidopsis thaliana; SB, Soybean.
| Target | Putative off-target locus | Off-target sequence | Mutation |
|---|---|---|---|
| Target site 1 | Chr1: Os01g0607900 (intron) | TACAAAAACTACCAGGCCAATGG | 0 |
| Chr5: Os05g05370 (intron) | AAGAATAACTACCAACCAAAAGA | 0 | |
| Chr6: Os06g0230000 (exon) | AAGAACACATACCAGCCTTATGT | 0 | |
| Chr11: 18470801(intergenic) | AAGAAGAACTACCACACCAAAAG | 0 | |
| Chr12: Os12g0586600 (exon) | AAGAACAAATACCACTGCAAGGG | 0 | |
| Target site 2 | Chr3: Os03g0403501 (exon) | AAGCTCGACAGGATGAAGGCCGC | 0 |
| Chr5: Os05g0124600 (intron) | AGGATCAACTGGAAGAAGCCAGG | 0 | |
| Chr6: 5669112 (intergenic) | AAGATCAATTGGGTAAAGACAGG | 0 | |
| Chr7: Os07g0119300 (exon) | AAGAGCAACTGGATGAGCACCGC | 0 | |
| Chr9: Os09g0134500 (exon) | AAGATCACCTGGATCAAGACCTT | 0 | |
| Target site 3 | Chr1: Os01g0830700 (exon) | CGACCTGTCCAAGGGCGAGTGGG | 0 |
| Chr2: Os02g0590800 (exon) | CGTCGAGTACAAGGACGGTTGGG | 0 | |
| Chr6: Os06g0611500 (exon) | CGGCCGGTACAAGGACGAGCGGG | 0 | |
| Chr9: 4319579 (intergenic) | CGAGCAGTACAGGGAGGATTGGG | 0 | |
| Chr10: 9307021 (intergenic) | CGATCAAAATAAGGACGCTTGGG | 0 |
Table S3 Identification of off-target effects.
| Target | Putative off-target locus | Off-target sequence | Mutation |
|---|---|---|---|
| Target site 1 | Chr1: Os01g0607900 (intron) | TACAAAAACTACCAGGCCAATGG | 0 |
| Chr5: Os05g05370 (intron) | AAGAATAACTACCAACCAAAAGA | 0 | |
| Chr6: Os06g0230000 (exon) | AAGAACACATACCAGCCTTATGT | 0 | |
| Chr11: 18470801(intergenic) | AAGAAGAACTACCACACCAAAAG | 0 | |
| Chr12: Os12g0586600 (exon) | AAGAACAAATACCACTGCAAGGG | 0 | |
| Target site 2 | Chr3: Os03g0403501 (exon) | AAGCTCGACAGGATGAAGGCCGC | 0 |
| Chr5: Os05g0124600 (intron) | AGGATCAACTGGAAGAAGCCAGG | 0 | |
| Chr6: 5669112 (intergenic) | AAGATCAATTGGGTAAAGACAGG | 0 | |
| Chr7: Os07g0119300 (exon) | AAGAGCAACTGGATGAGCACCGC | 0 | |
| Chr9: Os09g0134500 (exon) | AAGATCACCTGGATCAAGACCTT | 0 | |
| Target site 3 | Chr1: Os01g0830700 (exon) | CGACCTGTCCAAGGGCGAGTGGG | 0 |
| Chr2: Os02g0590800 (exon) | CGTCGAGTACAAGGACGGTTGGG | 0 | |
| Chr6: Os06g0611500 (exon) | CGGCCGGTACAAGGACGAGCGGG | 0 | |
| Chr9: 4319579 (intergenic) | CGAGCAGTACAGGGAGGATTGGG | 0 | |
| Chr10: 9307021 (intergenic) | CGATCAAAATAAGGACGCTTGGG | 0 |
| Variety | Grain length (mm) | Grain width (mm) | Grain thickness (mm) | 1000-grain weight (g) |
|---|---|---|---|---|
| YK17 | 7.29 ± 0.10 | 2.62 ± 0.05 | 2.10 ± 0.06 | 16.28 ± 1.16 |
| MT1 | 7.46 ± 0.05* | 2.82 ± 0.04* | 2.27 ± 0.07* | 18.20 ± 0.18* |
| MT2 | 7.52 ± 0.14* | 2.76 ± 0.06* | 2.19 ± 0.09* | 15.97 ± 0.60 |
| MT3 | 7.52 ± 0.02* | 2.79 ± 0.04* | 2.12 ± 0.07 | 15.10 ± 1.31 |
Table S4 Agronomic traits of YK17 and mutants (MT1, MT2 and MT3).
| Variety | Grain length (mm) | Grain width (mm) | Grain thickness (mm) | 1000-grain weight (g) |
|---|---|---|---|---|
| YK17 | 7.29 ± 0.10 | 2.62 ± 0.05 | 2.10 ± 0.06 | 16.28 ± 1.16 |
| MT1 | 7.46 ± 0.05* | 2.82 ± 0.04* | 2.27 ± 0.07* | 18.20 ± 0.18* |
| MT2 | 7.52 ± 0.14* | 2.76 ± 0.06* | 2.19 ± 0.09* | 15.97 ± 0.60 |
| MT3 | 7.52 ± 0.02* | 2.79 ± 0.04* | 2.12 ± 0.07 | 15.10 ± 1.31 |
| [1] | Barman H N, Sheng Z H, Fiaz S, Zhong M, Wu Y W, Cai Y C, Wang W, Jiao G A, Tang S Q, Wei X J, Hu P S. 2019. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol, 19(1): 109. |
| [2] | Biselli C, Cavalluzzo D, Perrini R, Gianinetti A, Bagnaresi P, Urso S, Orasen G, Desiderio F, Lupotto E, Cattivelli L, Valè G. 2014. Improvement of marker-based predictability of apparent amylose content in japonica rice through GBSS I allele mining. Rice, 7(1): 1. |
| [3] | Cai X L, Wang Z Y, Xing Y Y, Zhang J L, Hong M M. 1998. Aberrant splicing of intron 1 leads to the heterogeneous 5 ' UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J, 14(4): 459-465. |
| [4] | Fei Y Y, Yang J, Wang F Q, Fan F J, Li W Q, Wang J, Xu Y, Zhu J Y, Zhong W G. 2019. Production of two elite glutinous rice varieties by editing Wx gene. Rice Sci, 26(2): 118-124. |
| [5] | Gaudelli N M, Komor A C, Rees H A, Packer M S, Badran A H, Bryson D I, Liu D R. 2017. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature, 551: 464-471. |
| [6] | Huang L C, Sreenivasulu N, Liu Q Q. 2020a. Waxy editing: Old meets new. Trends Plant Sci, 25(10): 963-966. |
| [7] | Huang L C, Li Q F, Zhang C Q, Chu R, Gu Z W, Tan H Y, Zhao D S, Fan X L, Liu Q Q. 2020b. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol J, 18(11): 2164-2166. |
| [8] | Juliano B O. 1998. Varietal impact on rice quality. Cereal Foods World, 43(4): 207-222. |
| [9] | Komor A C, Kim Y B, Packer M S, Zuris J A, Liu D R. 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 533: 420-424. |
| [10] | Larkin P D, Park W D. 2003. Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice(Oryza sativa L.). Mol Breeding, 12: 335-339. |
| [11] | Li C, Zong Y, Wang Y P, Jin S, Zhang D B, Song Q N, Zhang R, Gao C X. 2018. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genom Biol, 19(1): 59. |
| [12] | Li H, Li X F, Xu Y, Liu H L, He M L, Tian X J, Wang Z Y, Wu X J, Bu Q Y, Yang J. 2020. High-efficiency reduction of rice amylose content via CRISPR/Cas9-mediated base editing. Rice Sci, 27(6): 445-448. |
| [13] | Liu L L, Ma X D, Liu S J, Zhu C L, Jiang L, Wang Y H, Shen Y, Ren Y, Dong H, Chen L M, Liu X, Zhao Z G, Zhai H Q, Wan J M. 2009. Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol Biol, 71: 609-626. |
| [14] | Mikami I, Uwatoko N, Ikeda Y, Yamaguchi J, Hirano H Y, Sujuki Y, Sano Y. 2008. Allelic diversification at the wx locus in landraces of Asian rice. Theor Appl Genet, 116(7): 979-989. |
| [15] | Molla K A, Yang Y N. 2019. CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol, 37(10): 1121-1142. |
| [16] | Monsur M B, Shao G N, Lv Y S, Ahmad S, Wei X J, Hu P S, Tang S Q. 2020. Base editing: The ever expanding clustered regularly interspaced short palindromic repeats (CRISPR) tool kit for precise genome editing in plants. Gene, 11(4): 466. |
| [17] | Sato H, Suzuki Y, Sakai M, Imbe T. 2002. Molecular characterization of wx-mq, a novel mutant gene for low-amylose content in endosperm of rice(Oryza sativa L.). Breeding Sci, 52(2): 131-135. |
| [18] | Teng B, Zeng R Z, Wang Y C, Liu Z Q, Zhang Z M, Zhu H T, Ding X H, Li W T, Zhang G Q. 2012. Detection of allelic variation at the Wx locus with single-segment substitution lines in rice(Oryza sativa L.). Mol Breeding, 30(1): 583-595. |
| [19] | Tian Z X, Qian Q, Liu Q Q, Yan M X, Liu X F, Yan C J, Liu G F, Gao Z Y, Tang S Z, Zeng D L, Wang Y H, Yu J M, Gu M H, Li J Y. 2009. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci USA, 106(51): 21760-21765. |
| [20] | Wanchana S, Toojinda T, Tragoonrung S, Vanavichit A. 2003. Duplicated coding sequence in the waxy allele of tropical glutinous rice(Oryza sativa L.). Plant Sci, 165(6): 1193-1199. |
| [21] | Wang Z Y, Zheng F Q, Shen G Z, Gao J P, Snusted D P, Li M G, Zhang J Z, Hong M M. 1995. Post-transcriptional regulation of the rice Waxy gene. Plant J, 7: 613-622. |
| [22] | Xu Y, Lin Q P, Li X F, Wang F Q, Chen Z H, Wang J, Li W Q, Fan F J, Tao Y J, Jiang Y J, Wei X D, Zhang R, Zhu Q H, Bu Q Y, Yang J, Gao C X. 2020. Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol J, 19(1): 11-13. |
| [23] | Yang J, Wang J, Fan F J, Zhu J Y, Chen T, Wang C L, Zheng T Q, Zhang J, Zhong W G, Xu J L. 2013. Development of AS-PCR marker based on a key mutation confirmed by resequencing of Wx-mp in milky princess and its application in japonica soft rice(Oryza sativa L.) breeding. Plant Breeding, 132(6): 595-603. |
| [24] | Zeng D C, Liu T L, Ma X L, Wang B, Zheng Z Y, Zhang Y L, Xie X R, Yang B W, Zhao Z, Zhu Q L, Liu Y G. 2020. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5'-UTR-intron editing improves grain quality in rice. Plant Biotechnol J, 18(12): 2385-2387. |
| [25] | Zhang C Q, Zhu J H, Chen S J, Fan X L, Li Q F, Lu Y, Wang M, Yu H X, Yi C D, Tang S Z, Gu M H, Liu Q Q. 2019. Wxlv, the ancestral allele of rice Waxy gene. Mol Plant, 12(8): 1157-1166. |
| [26] | Zhang C Q, Yang Y, Chen S J, Liu X J, Zhu J H, Lu Y, Li Q F, Fan X L, Tang S Z, Gu M H, Liu Q Q. 2021. A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J Integr Plant Biol, 63(5): 889-901. |
| [27] | Zhang J S, Zhang H, Botella J R, Zhu J K. 2018. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J Integr Plant Biol, 60(5): 369-375. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||