
Rice Science ›› 2020, Vol. 27 ›› Issue (2): 113-123.DOI: 10.1016/j.rsci.2020.01.002
收稿日期:2018-09-18
接受日期:2019-01-15
出版日期:2020-03-28
发布日期:2019-11-28
. [J]. Rice Science, 2020, 27(2): 113-123.
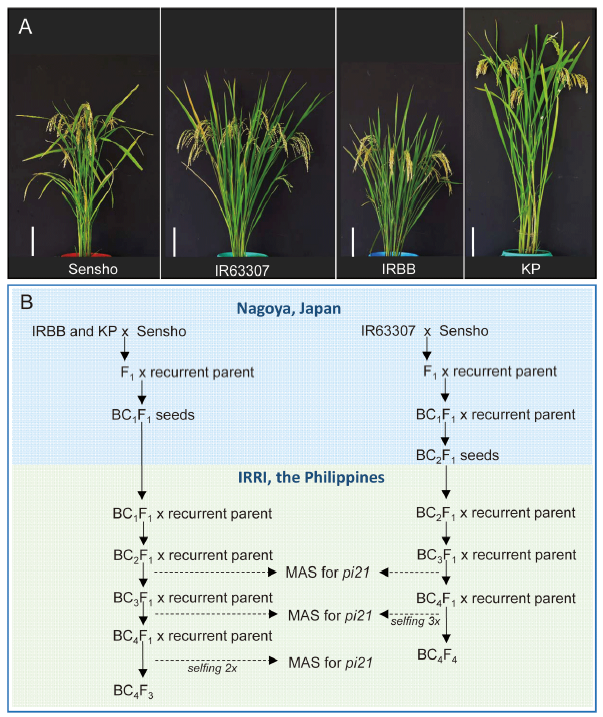
Fig. 1. Introgression of pi21 allele from Sensho to different rice genetic backgrounds. A, Gross morphology of Sensho and the recipient parents IR63307, IRBB and KP. Bar = 20 cm. B, Marker-assisted breeding scheme used to transfer the pi21 allele from Sensho to the different rice backgrounds. The blue panel indicates the initial crosses and backcrosses that were carried out in Nagoya, Japan, whereas the green panel indicates the marker-assisted backcrossing and generation advance that were conducted at International Rice Research Institute (IRRI) in the Philippines. MAS, Marker-assisted sclection.
| Isolate name | Groupb | Subgroupc | Philippine collection site |
|---|---|---|---|
| Ca89 | I | b | Luzon and Mindanao islands |
| IK81-3 | II | - | Southern Luzon island |
| M39-1-3-8-1 | III | a | Southern Luzon island |
| V850256 | III | c | Southern Luzon island |
| M64-1-3-9-1 | III | a | Southern Luzon island |
| V86010 | IV | e | Central and Southern Luzon island |
| BZ64-1 | IV | - | unknown |
| BN111 | V | a | Southern Luzon island |
| PO6-6 | V | b | Southern Luzon island |
| M39-1-2-21-2 | VI | b | Central and Southern Luzon island |
| JMB8401 | XII | a | Southern Luzon island |
| 43 | XVIII | - | unknown |
| M36-1-3-10-1 | XIX | b | Southern Luzon and Mindanao islands |
| 9475-1-3 | XX | - | unknown |
| BN209 | XXII | a | Southern Luzon island |
| M101-1-2-9-1 | XXII | a | Southern Luzon island |
| IK81-25 | XXIII | - | unknown |
| JMB840610 | XXIV | b | Southern Luzon island |
| 5167-1 | XIX | - | unknown |
| Pi9-G7-2K-1 | - | - | - |
Supplemental Table 1. Twenty blast isolates from the Philippines used in leaf blast isolate-specific screening of WISH lines carrying the pi21 allele.a
| Isolate name | Groupb | Subgroupc | Philippine collection site |
|---|---|---|---|
| Ca89 | I | b | Luzon and Mindanao islands |
| IK81-3 | II | - | Southern Luzon island |
| M39-1-3-8-1 | III | a | Southern Luzon island |
| V850256 | III | c | Southern Luzon island |
| M64-1-3-9-1 | III | a | Southern Luzon island |
| V86010 | IV | e | Central and Southern Luzon island |
| BZ64-1 | IV | - | unknown |
| BN111 | V | a | Southern Luzon island |
| PO6-6 | V | b | Southern Luzon island |
| M39-1-2-21-2 | VI | b | Central and Southern Luzon island |
| JMB8401 | XII | a | Southern Luzon island |
| 43 | XVIII | - | unknown |
| M36-1-3-10-1 | XIX | b | Southern Luzon and Mindanao islands |
| 9475-1-3 | XX | - | unknown |
| BN209 | XXII | a | Southern Luzon island |
| M101-1-2-9-1 | XXII | a | Southern Luzon island |
| IK81-25 | XXIII | - | unknown |
| JMB840610 | XXIV | b | Southern Luzon island |
| 5167-1 | XIX | - | unknown |
| Pi9-G7-2K-1 | - | - | - |
| Cultivar and line | Initial reading | Final reading | |||||
|---|---|---|---|---|---|---|---|
| SES | DLA (%) | Phenotype | SES | DLA (%) | Phenotype | ||
| Lijiangxintuanheigu (LTH, susceptible control) | 5.3 | 34 | MR | ND | ND | S | |
| CO39 (susceptible control) | 7.1 | 60 | S | 8.2 | 80 | S | |
| IR65482-4-136-2-2 (resistant control) | 1.4 | 1 | R | 1.6 | 1 | R | |
| Sensho | 1.1 | 2 | R | 1.5 | 2 | R | |
| IR63307-4B0-B-2 (IR63307) | 0.5 | 1 | R | 0.5 | 1 | R | |
| WISH110:1-1-11-12-1 | 1.0 | 1 | R | 1.0 | 1 | R | |
| WISH110:1-1-11-5-4 | 2.0 | 1 | R | 0.5 | 1 | R | |
| WISH110:2-5-2-1-1 | 1.0 | 1 | R | 0.5 | 1 | R | |
| WISH110:2-5-2-12-1 | 0.0 | 0 | R | 0.5 | 1 | R | |
| WISH110:2-5-2-22-1 | 0.5 | 1 | R | 0.5 | 1 | R | |
| IRBB4/5/13/21 (IRBB) | 4.0 | 5 | MR | 5.0 | 15 | MR | |
| WISH48:1-3-1-1 | 2.0 | 1 | R | 2.0 | 1 | R | |
| WISH48:1-3-18-1 | 2.0 | 3 | R | 3.5 | 5 | R | |
| WISH48:1-3-2-1 | 1.0 | 1 | R | 1.0 | 1 | R | |
| WISH48:1-3-20-1 | 3.0 | 3 | R | 1.0 | 1 | R | |
| WISH48:1-3-21-1 | 1.0 | 1 | R | 2.0 | 1 | R | |
| Kinandang Patong (KP) | 0.0 | 0 | R | 0.5 | 0 | R | |
| WISH40:1-3-16-11 | 0.0 | 0 | R | 0.5 | 0 | R | |
| WISH40:1-3-3-9 | 0.0 | 0 | R | 0.0 | 0 | R | |
| WISH40:1-3-7-7 | 0.0 | 0 | R | 0.0 | 0 | R | |
Table 1 Reaction of WISH lines to natural leaf blast infection in the field based on SES scores (IRRI, 2014).
| Cultivar and line | Initial reading | Final reading | |||||
|---|---|---|---|---|---|---|---|
| SES | DLA (%) | Phenotype | SES | DLA (%) | Phenotype | ||
| Lijiangxintuanheigu (LTH, susceptible control) | 5.3 | 34 | MR | ND | ND | S | |
| CO39 (susceptible control) | 7.1 | 60 | S | 8.2 | 80 | S | |
| IR65482-4-136-2-2 (resistant control) | 1.4 | 1 | R | 1.6 | 1 | R | |
| Sensho | 1.1 | 2 | R | 1.5 | 2 | R | |
| IR63307-4B0-B-2 (IR63307) | 0.5 | 1 | R | 0.5 | 1 | R | |
| WISH110:1-1-11-12-1 | 1.0 | 1 | R | 1.0 | 1 | R | |
| WISH110:1-1-11-5-4 | 2.0 | 1 | R | 0.5 | 1 | R | |
| WISH110:2-5-2-1-1 | 1.0 | 1 | R | 0.5 | 1 | R | |
| WISH110:2-5-2-12-1 | 0.0 | 0 | R | 0.5 | 1 | R | |
| WISH110:2-5-2-22-1 | 0.5 | 1 | R | 0.5 | 1 | R | |
| IRBB4/5/13/21 (IRBB) | 4.0 | 5 | MR | 5.0 | 15 | MR | |
| WISH48:1-3-1-1 | 2.0 | 1 | R | 2.0 | 1 | R | |
| WISH48:1-3-18-1 | 2.0 | 3 | R | 3.5 | 5 | R | |
| WISH48:1-3-2-1 | 1.0 | 1 | R | 1.0 | 1 | R | |
| WISH48:1-3-20-1 | 3.0 | 3 | R | 1.0 | 1 | R | |
| WISH48:1-3-21-1 | 1.0 | 1 | R | 2.0 | 1 | R | |
| Kinandang Patong (KP) | 0.0 | 0 | R | 0.5 | 0 | R | |
| WISH40:1-3-16-11 | 0.0 | 0 | R | 0.5 | 0 | R | |
| WISH40:1-3-3-9 | 0.0 | 0 | R | 0.0 | 0 | R | |
| WISH40:1-3-7-7 | 0.0 | 0 | R | 0.0 | 0 | R | |
| Line name | Blast isolates | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
| LTH | 3b/S | 3/S | 0/R | 3/S | 4/S | 4/S | 3/S | 4/S | 3/S | 3/S | 5/S | 5/S | 3/S | 5/S | 5/S | 5/S | 5/S | 5/S | 4/S | 4/S |
| CO39 | 4/S | 3/S | 2/R | 4/S | 4/S | 4/S | 3/S | 4/S | 4/S | 3/S | 5/S | 5/S | 4/S | 4/S | 4/S | 5/S | 5/S | 5/S | 4/S | 0/R |
| IR65482-4-136-2-2 | 0/R | 0/R | 0/R | 0/R | 0/R | 0/R | 0/R | 0/R | 0/R | 0/R | 1/R | 0/R | 4/S | 0/R | 0/R | 1/R | 2/R | 0/R | 0/R | 0/R |
| IR72 | 0/R | 0/R | 0/R | 1/R | 0/R | 3/S | 0/R | 0/R | 0/R | 0/R | 2/R | 1/R | 0/R | 0/R | 0/R | 0/R | 2/R | 1/R | 0/R | 2/R |
| Sensho | 0/R | 1/R | 0/R | 4/S | 2/R | 1/R | 1/R | 1/R | 3/S | 3/S | 2/R | 2/R | 1/R | 0/R | 2/R | 4/S | 2/R | 1/R | 1/R | 0/R |
| IR63307 | 2/R | 1/R | 0/R | 0/R | 0/R | 2/R | 1/R | 2/R | 2/R | 1/R | 1/R | 2/R | 2/R | 0/R | 5/S | 5/S | 2/R | 0/R | 4/S | 0/R |
| WISH 110:1-1-11-12-1 | 1/R | 0/R | 0/R | 0/R | 0/R | 1/R | 1/R | 0/R | 1/R | 0/R | 1/R | 1/R | 1/R | 0/R | 2/R | 2/R | 1/R | 0/R | 2/R | 0/R |
| WISH 110:1-1-11-5-4 | 1/R | 1/R | 0/R | 0/R | 0/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 0/R | 2/R | 2/R | 1/R | 0/R | 1/R | 0/R |
| WISH 110:2-5-2-1-1 | 1/R | 0/R | 0/R | 0/R | 0/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 0/R | 2/R | 2/R | 1/R | 0/R | 2/R | 0/R |
| WISH 110:2-5-2-12-1 | 1/R | 0/R | 0/R | 1/R | 0/R | 1/R | 0/R | 1/R | 1/R | 0/R | 1/R | 1/R | 1/R | 0/R | 1/R | 1/R | 1/R | 0/R | 2/R | 0/R |
| WISH 110:2-5-2-22-1 | 1/R | 1/R | 0/R | 0/R | 0/R | 1/R | 0/R | 1/R | 1/R | 0/R | 1/R | 1/R | 1/R | 0/R | 2/R | 2/R | 1/R | 0/R | 2/R | 0/R |
| IRBB | 2/R | 3/S | 1/R | 4/S | 0/R | 2/R | 3/S | 2/R | 0/R | 0/R | 5/S | 0/R | 4/S | 4/S | 2/R | 4/S | 5/S | 5/S | 0/R | 0/R |
| WISH 48:1-3-1-1 | 1/R | 1/R | 0/R | 3/S | 0/R | 1/R | 1/R | 0/R | 0/R | 0/R | 2/R | 0/R | 1/R | 2/R | 1/R | 4/S | 4/S | 2/R | 0/R | 0/R |
| WISH 48:1-3-18-1 | 1/R | 1/R | 0/R | 3/S | 0/R | 2/R | 2/R | 1/R | 0/R | 0/R | 2/R | 0/R | 1/R | 2/R | 1/R | 4/S | 4/S | 3/S | 1/R | 0/R |
| WISH 48:1-3-2-1 | 1/R | 1/R | 1/R | 1/R | 0/R | 0/R | 1/R | 1/R | 0/R | 0/R | 2/R | 0/R | 1/R | 1/R | 0/R | 3/S | 3/S | 2/R | 0/R | 0/R |
| WISH 48:1-3-20-1 | 1/R | 2/R | 1/R | 4/S | 0/R | 2/R | 1/R | 1/R | 0/R | 0/R | 3/S | 0/R | 1/R | 1/R | 1/R | 4/S | 4/S | 3/S | 0/R | 0/R |
| WISH 48:1-3-21-1 | 1/R | 1/R | 0/R | 3/S | ND | 0/R | 0/R | 1/R | 0/R | 0/R | 1/R | 0/R | 2/R | 1/R | 1/R | 4/S | 4/S | 2/R | 0/R | 0/R |
| KP | 1/R | 2/R | 0/R | 3/S | 4/S | 1/R | 1/R | 2/R | 4/S | 3/S | 3/S | 5/S | 1/R | 2/R | 4/S | 2/R | 3/S | 1/R | 4/S | 0/R |
| WISH 40:1-3-16-11 | 1/R | 0/R | 0/R | 2/R | 2/R | 1/R | 0/R | 1/R | 1/R | 0/R | 2/R | 2/R | 0/R | 1/R | 2/R | 1/R | 2/R | 1/R | 3/S | 1/R |
| WISH 40:1-3-3-9 | 1/R | 1/R | 0/R | 2/R | 2/R | 1/R | 0/R | 1/R | 1/R | 2/R | 1/R | 2/R | 0/R | 1/R | 2/R | 1/R | 1/R | 1/R | 1/R | 0/R |
| WISH 40:1-3-7-7 | 1/R | 0/R | 0/R | 1/R | 0/R | 1/R | 1/R | 0/R | 2/R | 1/R | 1/R | 2/R | 0/R | 1/R | 2/R | 1/R | 1/R | 1/R | 1/R | 0/R |
Supplemental Table 2. Reaction of WISH lines to twenty leaf blast isolates in the glasshouse.
| Line name | Blast isolates | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
| LTH | 3b/S | 3/S | 0/R | 3/S | 4/S | 4/S | 3/S | 4/S | 3/S | 3/S | 5/S | 5/S | 3/S | 5/S | 5/S | 5/S | 5/S | 5/S | 4/S | 4/S |
| CO39 | 4/S | 3/S | 2/R | 4/S | 4/S | 4/S | 3/S | 4/S | 4/S | 3/S | 5/S | 5/S | 4/S | 4/S | 4/S | 5/S | 5/S | 5/S | 4/S | 0/R |
| IR65482-4-136-2-2 | 0/R | 0/R | 0/R | 0/R | 0/R | 0/R | 0/R | 0/R | 0/R | 0/R | 1/R | 0/R | 4/S | 0/R | 0/R | 1/R | 2/R | 0/R | 0/R | 0/R |
| IR72 | 0/R | 0/R | 0/R | 1/R | 0/R | 3/S | 0/R | 0/R | 0/R | 0/R | 2/R | 1/R | 0/R | 0/R | 0/R | 0/R | 2/R | 1/R | 0/R | 2/R |
| Sensho | 0/R | 1/R | 0/R | 4/S | 2/R | 1/R | 1/R | 1/R | 3/S | 3/S | 2/R | 2/R | 1/R | 0/R | 2/R | 4/S | 2/R | 1/R | 1/R | 0/R |
| IR63307 | 2/R | 1/R | 0/R | 0/R | 0/R | 2/R | 1/R | 2/R | 2/R | 1/R | 1/R | 2/R | 2/R | 0/R | 5/S | 5/S | 2/R | 0/R | 4/S | 0/R |
| WISH 110:1-1-11-12-1 | 1/R | 0/R | 0/R | 0/R | 0/R | 1/R | 1/R | 0/R | 1/R | 0/R | 1/R | 1/R | 1/R | 0/R | 2/R | 2/R | 1/R | 0/R | 2/R | 0/R |
| WISH 110:1-1-11-5-4 | 1/R | 1/R | 0/R | 0/R | 0/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 0/R | 2/R | 2/R | 1/R | 0/R | 1/R | 0/R |
| WISH 110:2-5-2-1-1 | 1/R | 0/R | 0/R | 0/R | 0/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 1/R | 0/R | 2/R | 2/R | 1/R | 0/R | 2/R | 0/R |
| WISH 110:2-5-2-12-1 | 1/R | 0/R | 0/R | 1/R | 0/R | 1/R | 0/R | 1/R | 1/R | 0/R | 1/R | 1/R | 1/R | 0/R | 1/R | 1/R | 1/R | 0/R | 2/R | 0/R |
| WISH 110:2-5-2-22-1 | 1/R | 1/R | 0/R | 0/R | 0/R | 1/R | 0/R | 1/R | 1/R | 0/R | 1/R | 1/R | 1/R | 0/R | 2/R | 2/R | 1/R | 0/R | 2/R | 0/R |
| IRBB | 2/R | 3/S | 1/R | 4/S | 0/R | 2/R | 3/S | 2/R | 0/R | 0/R | 5/S | 0/R | 4/S | 4/S | 2/R | 4/S | 5/S | 5/S | 0/R | 0/R |
| WISH 48:1-3-1-1 | 1/R | 1/R | 0/R | 3/S | 0/R | 1/R | 1/R | 0/R | 0/R | 0/R | 2/R | 0/R | 1/R | 2/R | 1/R | 4/S | 4/S | 2/R | 0/R | 0/R |
| WISH 48:1-3-18-1 | 1/R | 1/R | 0/R | 3/S | 0/R | 2/R | 2/R | 1/R | 0/R | 0/R | 2/R | 0/R | 1/R | 2/R | 1/R | 4/S | 4/S | 3/S | 1/R | 0/R |
| WISH 48:1-3-2-1 | 1/R | 1/R | 1/R | 1/R | 0/R | 0/R | 1/R | 1/R | 0/R | 0/R | 2/R | 0/R | 1/R | 1/R | 0/R | 3/S | 3/S | 2/R | 0/R | 0/R |
| WISH 48:1-3-20-1 | 1/R | 2/R | 1/R | 4/S | 0/R | 2/R | 1/R | 1/R | 0/R | 0/R | 3/S | 0/R | 1/R | 1/R | 1/R | 4/S | 4/S | 3/S | 0/R | 0/R |
| WISH 48:1-3-21-1 | 1/R | 1/R | 0/R | 3/S | ND | 0/R | 0/R | 1/R | 0/R | 0/R | 1/R | 0/R | 2/R | 1/R | 1/R | 4/S | 4/S | 2/R | 0/R | 0/R |
| KP | 1/R | 2/R | 0/R | 3/S | 4/S | 1/R | 1/R | 2/R | 4/S | 3/S | 3/S | 5/S | 1/R | 2/R | 4/S | 2/R | 3/S | 1/R | 4/S | 0/R |
| WISH 40:1-3-16-11 | 1/R | 0/R | 0/R | 2/R | 2/R | 1/R | 0/R | 1/R | 1/R | 0/R | 2/R | 2/R | 0/R | 1/R | 2/R | 1/R | 2/R | 1/R | 3/S | 1/R |
| WISH 40:1-3-3-9 | 1/R | 1/R | 0/R | 2/R | 2/R | 1/R | 0/R | 1/R | 1/R | 2/R | 1/R | 2/R | 0/R | 1/R | 2/R | 1/R | 1/R | 1/R | 1/R | 0/R |
| WISH 40:1-3-7-7 | 1/R | 0/R | 0/R | 1/R | 0/R | 1/R | 1/R | 0/R | 2/R | 1/R | 1/R | 2/R | 0/R | 1/R | 2/R | 1/R | 1/R | 1/R | 1/R | 0/R |
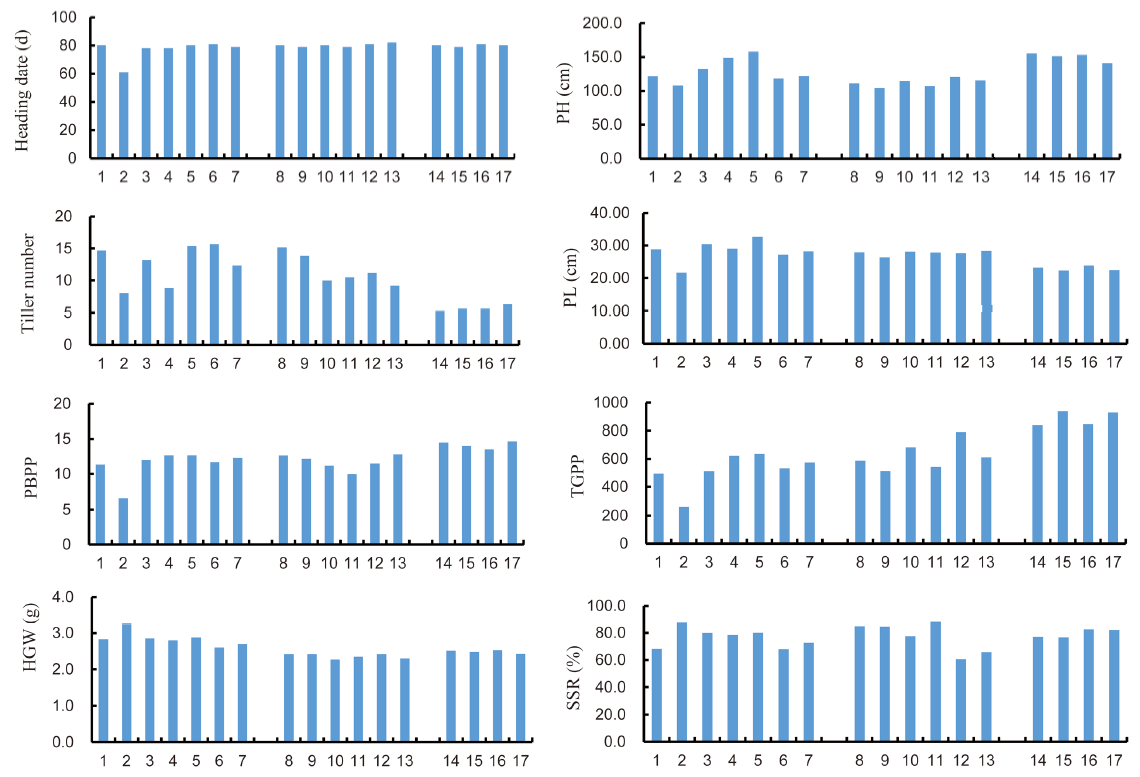
Fig. 2. Morphometric data on agronomic performance of Wonder Rice Initiative for Food Security and Health (WISH) lines in different backgrounds. 1, IR63307; 2, Sensho; 3, WISH110:1-1-11-12-1; 4, WISH110:1-1-11-5-4; 5, WISH110:2-5-2-1-1; 6, WISH110:2-5-2-12-1; 7, WISH110:2-5-2-22-1; 8, IRBB; 9, WISH48:1-3-1-1; 10, WISH48:1-3-2-1; 11, WISH48:1-3-18-1; 12, WISH48:1-3-20-1; 13, WISH48:1-3-21-1; 14, KP; 15, WISH40:1-3-16-11; 16, WISH40:1-3-3-9; 17, WISH40:1-3-7-7; PH, Plant height; PL, Panicle length; PBPP, Number of primary branches per panicle; TGPP, Total grain number per panicle; HGW, 100-grain weight; SSR, Seed-setting rate.
| Line name | Raw grain length (mm) | Raw grain width (mm) | Ratio of | Cooked grain length (mm) | Cooked grain width (mm) | Ratio of cooked grain length and width | Chalkiness (%) | Amylose content | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| grain length and width | ||||||||||||||||
| DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | |
| Sensho | 6 | 6.1 | 2.9 | 2.8 | 2.1 | 2.2 | 10 | 9.9 | 4.3 | 4.4 | 2.4 | 2.3 | 6.8 | 16.4 | 21 | 17.4 |
| IR63307 | 5.8 | 5.9 | 2.8 | 2.7 | 2.1 | 2.2 | 9.2 | 8.9 | 4 | 3.9 | 2.3 | 2.3 | 7.3*a | 10.5*a | 24* | 21.1* |
| WISH 110:1-1-11-12-1 | 5.9 | 6 | 2.8 | 2.8 | 2.1 | 2.1 | 9.2 | 9.1 | 4.1 | 3.9 | 2.2 | 2.3 | 3.5*b | 8.3*b | 23* | 20.5* |
| WISH 110:1-1-11-5-4 | 5.9 | 5.9 | 2.8 | 2.7 | 2.1 | 2.2 | 9.5 | 9.4 | 4 | 3.9 | 2.4 | 2.4 | 1.6*c | 6.2*c | 23* | 22.4* |
| WISH 110:2-5-2-1-1 | 6.1 | 6 | 2.8 | 2.7 | 2.2 | 2.2 | 10 | 9.7 | 4.1 | 3.9 | 2.5 | 2.5 | 0.1*d | 3.2*d | 23* | 22.7* |
| WISH 110:2-5-2-12-1 | 5.7 | 5.6 | 2.8 | 2.7 | 2 | 2.1 | 9.1 | 8.8 | 4.1 | 3.8 | 2.2 | 2.3 | 1.7*c | 4.7*e | 23* | 21.6* |
| WISH 110:2-5-2-22-1 | 5.5 | 5.5 | 2.8 | 2.8 | 2 | 2 | 9.3 | 8.9 | 4.2 | 4 | 2.2 | 2.2 | 5.4*e | 6.9*f | 23* | 22.0* |
| IRBB | 6.9 | 6.8 | 2.2 | 2 | 3.1 | 3.4 | 10 | 10.3 | 4 | 3.5 | 2.6 | 2.9 | 0.3a | 0.4a | 13*a | 12.9*a |
| WISH 48:1-3-1-1 | 6.6 | 6.6 | 2.3 | 2.1 | 2.9 | 3.1 | 10 | 10.2 | 3.5 | 3.5 | 2.9 | 2.9 | 0.3a | 0.8b | 19*b | 15.3*b |
| WISH 48:1-3-18-1 | 6.4 | 6.8 | 2.3 | 2.1 | 2.8 | 3.2 | 11 | 10.5 | 3.5 | 3.2 | 3 | 3.3 | 1.2b | 0.2c | 21*c | 18.2*c |
| WISH 48:1-3-2-1 | 6.3 | 6.5 | 2.3 | 2.1 | 2.7 | 3.1 | 9.9 | 10.1 | 3.6 | 3.4 | 2.8 | 3 | 1.6c | 4.5d | 21*c | 19.6*d |
| WISH 48:1-3-20-1 | 6.7 | 6.9 | 2.3 | 2.1 | 2.9 | 3.3 | 9.9 | 10.4 | 3.6 | 3.5 | 2.8 | 3 | 5.6d | 7.9e | 20*d | 17.0*e |
| WISH 48:1-3-21-1 | 6.5 | 6.7 | 2.3 | 2.1 | 2.8 | 3.2 | 9.8 | 9.9 | 3.5 | 3.4 | 2.8 | 2.9 | 0.6e | 2.0f | 18*b | 18.9*f |
| KP | 5.1 | 5 | 2.9 | 2.7 | 1.8 | 1.9 | 8.7 | 8 | 4 | 3.8 | 2.2 | 2.1 | 7.5a | 8.5a | 23* | 21.3* |
| WISH 40:1-3-16-11 | 5 | 5.2 | 2.9 | 2.9 | 1.7 | 1.8 | 8.5 | 8.7 | 3.8 | 3.9 | 2.2 | 2.2 | 17.0b | 8.4a | 23* | 21.2* |
| WISH 40:1-3-3-9 | 5 | 5 | 2.9 | 2.8 | 1.7 | 1.8 | 8.4 | 8.3 | 3.9 | 3.8 | 2.2 | 2.2 | 10.2c | 14.5b | 24* | 22.0* |
| WISH 40:1-3-7-7 | 5.1 | 5 | 3 | 2.8 | 1.7 | 1.8 | 9.5 | 8.7 | 4.2 | 4 | 2.3 | 2.2 | 6.60d | 13.1c | 22* | 21.6* |
Supplemental Table 3. Grain quality of WISH lines having the pi21 allele for leaf blast resistance from Sensho measured during the dry (DS) and wet season (WS) of 2017.
| Line name | Raw grain length (mm) | Raw grain width (mm) | Ratio of | Cooked grain length (mm) | Cooked grain width (mm) | Ratio of cooked grain length and width | Chalkiness (%) | Amylose content | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| grain length and width | ||||||||||||||||
| DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | |
| Sensho | 6 | 6.1 | 2.9 | 2.8 | 2.1 | 2.2 | 10 | 9.9 | 4.3 | 4.4 | 2.4 | 2.3 | 6.8 | 16.4 | 21 | 17.4 |
| IR63307 | 5.8 | 5.9 | 2.8 | 2.7 | 2.1 | 2.2 | 9.2 | 8.9 | 4 | 3.9 | 2.3 | 2.3 | 7.3*a | 10.5*a | 24* | 21.1* |
| WISH 110:1-1-11-12-1 | 5.9 | 6 | 2.8 | 2.8 | 2.1 | 2.1 | 9.2 | 9.1 | 4.1 | 3.9 | 2.2 | 2.3 | 3.5*b | 8.3*b | 23* | 20.5* |
| WISH 110:1-1-11-5-4 | 5.9 | 5.9 | 2.8 | 2.7 | 2.1 | 2.2 | 9.5 | 9.4 | 4 | 3.9 | 2.4 | 2.4 | 1.6*c | 6.2*c | 23* | 22.4* |
| WISH 110:2-5-2-1-1 | 6.1 | 6 | 2.8 | 2.7 | 2.2 | 2.2 | 10 | 9.7 | 4.1 | 3.9 | 2.5 | 2.5 | 0.1*d | 3.2*d | 23* | 22.7* |
| WISH 110:2-5-2-12-1 | 5.7 | 5.6 | 2.8 | 2.7 | 2 | 2.1 | 9.1 | 8.8 | 4.1 | 3.8 | 2.2 | 2.3 | 1.7*c | 4.7*e | 23* | 21.6* |
| WISH 110:2-5-2-22-1 | 5.5 | 5.5 | 2.8 | 2.8 | 2 | 2 | 9.3 | 8.9 | 4.2 | 4 | 2.2 | 2.2 | 5.4*e | 6.9*f | 23* | 22.0* |
| IRBB | 6.9 | 6.8 | 2.2 | 2 | 3.1 | 3.4 | 10 | 10.3 | 4 | 3.5 | 2.6 | 2.9 | 0.3a | 0.4a | 13*a | 12.9*a |
| WISH 48:1-3-1-1 | 6.6 | 6.6 | 2.3 | 2.1 | 2.9 | 3.1 | 10 | 10.2 | 3.5 | 3.5 | 2.9 | 2.9 | 0.3a | 0.8b | 19*b | 15.3*b |
| WISH 48:1-3-18-1 | 6.4 | 6.8 | 2.3 | 2.1 | 2.8 | 3.2 | 11 | 10.5 | 3.5 | 3.2 | 3 | 3.3 | 1.2b | 0.2c | 21*c | 18.2*c |
| WISH 48:1-3-2-1 | 6.3 | 6.5 | 2.3 | 2.1 | 2.7 | 3.1 | 9.9 | 10.1 | 3.6 | 3.4 | 2.8 | 3 | 1.6c | 4.5d | 21*c | 19.6*d |
| WISH 48:1-3-20-1 | 6.7 | 6.9 | 2.3 | 2.1 | 2.9 | 3.3 | 9.9 | 10.4 | 3.6 | 3.5 | 2.8 | 3 | 5.6d | 7.9e | 20*d | 17.0*e |
| WISH 48:1-3-21-1 | 6.5 | 6.7 | 2.3 | 2.1 | 2.8 | 3.2 | 9.8 | 9.9 | 3.5 | 3.4 | 2.8 | 2.9 | 0.6e | 2.0f | 18*b | 18.9*f |
| KP | 5.1 | 5 | 2.9 | 2.7 | 1.8 | 1.9 | 8.7 | 8 | 4 | 3.8 | 2.2 | 2.1 | 7.5a | 8.5a | 23* | 21.3* |
| WISH 40:1-3-16-11 | 5 | 5.2 | 2.9 | 2.9 | 1.7 | 1.8 | 8.5 | 8.7 | 3.8 | 3.9 | 2.2 | 2.2 | 17.0b | 8.4a | 23* | 21.2* |
| WISH 40:1-3-3-9 | 5 | 5 | 2.9 | 2.8 | 1.7 | 1.8 | 8.4 | 8.3 | 3.9 | 3.8 | 2.2 | 2.2 | 10.2c | 14.5b | 24* | 22.0* |
| WISH 40:1-3-7-7 | 5.1 | 5 | 3 | 2.8 | 1.7 | 1.8 | 9.5 | 8.7 | 4.2 | 4 | 2.3 | 2.2 | 6.60d | 13.1c | 22* | 21.6* |
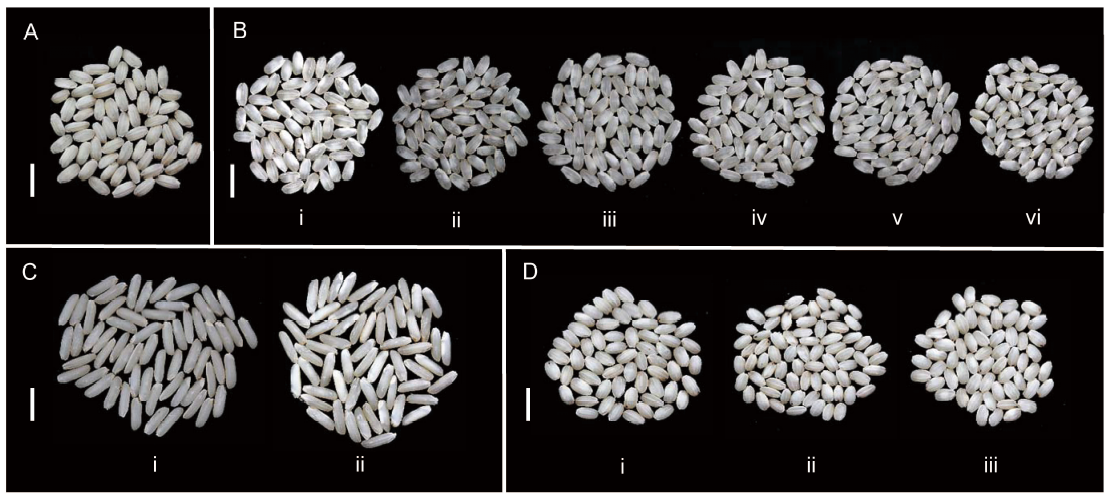
Fig. 3. Variation in the overall chalkiness degree of Wonder Rice Initiative for Food Security and Health (WISH) lines in different genetic backgrounds. A, Grains of Sensho. B, Grains of IR63307 (i), WISH110:2-5-2-1-1 (ii), WISH110:1-1-11-5-4 (iii), WISH110:2-5-2-12-1 (iv), WISH110:1-1-11-12-1 (v) and WISH110:2-5-2-22-1 (vi) in the background of IR63307, showing significantly lower chalkiness degrees compared to the recurrent parent. C, Grains of IRBB (i) and WISH48:1-3-20-2 (ii) in the IRBB background, showing a significantly higher chalkiness degree compared to the recurrent parent. D, Grains of KP (i), WISH40:1-3-3-9 (ii) and WISH40:1-3-16-11 (iii) in the KP background, showing higher chalkiness degrees compared to the recurrent parent. Bars = 10 mm.
| [1] | Bonman J M, Vergel De Dios T I, Khin M M. 1986. Physiological specialization of Pyricularia oryzae in the Philippines. Plant Dis, 70: 767-769. |
| [2] | Chen H, Iqbal M, Yang R C, Spanner D. 2016. Effect of Lr34/Yr18 on agronomic and quality traits in spring wheat mapping population and implications for breeding. Mol Breeding, 36: 53. |
| [3] | Cherif M, Harrabi M. 1993. Transgressive segregation for resistance to Pyrenophora teres in barley. Plant Pathol, 42(4): 617-621. |
| [4] | Dean R A, Talbot N J, Ebbole D J, Farman M L, Mitchell T K, Orbach M J, Thon M, Kulkarni R, Xu J R, Pan H Q, Read N D, Lee Y H, Carbone I, Brown D, Oh Y Y, Donofrio N, Jeong J S, Soanes D M, Djonovic S, Kolomiets E, Rehmeyer C, Li W X, Harding M, Kim S, Lebrun M H, Bohnert H, Coughlan S, Butler J, Calvo S, Ma L J, Nicol R, Purcell S, Nusbaum C, Galagan J E, Birren B W. 2005. The genome, sequence of the rice blast fungus Magnaporthe grisea. Nature, 434: 980-986. |
| [5] | de Vicente M C, Tanksley S D. 1993. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics, 134: 585-596. |
| [6] | Fujita D, Ebron L A, Kobayashi N, Fukuta Y. 2009. DNA marker analysis of blast resistance Pib and Pita in IRRI-bred rice varieties comparing with gene estimation by a differential system. In: Wang G L, Valent B. Advances in Genetics, Genomics and Control of Rice. Netherlands: Springer: 315-324. |
| [7] | Fukuoka S, Okuno K. 2001. QTL analysis and mapping of pi21, a recessive gene for field resistance to rice blast in Japanese upland rice. Theor Appl Genet, 103: 185-190. |
| [8] | Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebaba K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M. 2009. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science, 325: 998-1001. |
| [9] | Fukuoka S, Mizobuchi R, Saka N, Ivan S, Matsumoto T, Okuno K, Yano M. 2012. A multiple gene complex on rice chromosome 4 is involved in durable resistance to rice blast. Theor Appl Genet, 125(3): 551-599. |
| [10] | Fukuoka S, Saka N, Mizukami Y, Koga H, Yamanouchi U, Yoshioka Y, Hayashi N, Ebana K, Mizobuchi R, Yano M. 2015. Gene pyramiding enhances durable blast disease resistance in rice. Sci Rep, 5: 7773. |
| [11] | Fukuta Y, Teleblanco-Yanoria M J, Imbe T, Tsunematsu H, Kato H, Ban T, Ebron L A, Hayashi N, Ando I, Khush G S. 2004. Monogenic lines as an international standard differential set for blast resistance in rice (Oryza sativa L.). Rice Genet Newsl, 21: 70. |
| [12] | Goto I. 1970. Genetic studies on the resistance of rice plant to the blast fungus: I. Inheritance of resistance in crosses Sensho × H-79 and Imochishirazu × H-79. Ann Phytopathol Soc Jpn, 36: 304-312. |
| [13] | Hayashi N, Kobayashi N, Vera Cruz C M, Fukuta Y. 2009. Protocols for sampling of disease specimens and evaluation of blast disease in rice Japan International Research Center for Agricultural Sciences (JIRCAS), Tsukuba, Japan. Work Rep, 6: 17-33. |
| [14] | Horo J T, Fuji T, Yamashita Y, McGoey S, Koizumi S. 2016. Rice blast control efficacy of three genes ( Pib, pi21 and Pb1) conferring complete and partial resistance. Jpn Agric Res Q, 50(3): 209-217. |
| [15] | Huang N, Angeles E R, Domingo J, Magpantay G, Singh S, Zhang G, Kumaravadivel N, Bennet J, Khush G S. 1997. Pyramiding of bacterial blight resistance genes in rice: Marker-assisted selection using RFLP and PCR. Theor Appl Genet, 96(3): 313-320. |
| [16] | Imbe T, Oba S, Yanoria M J T, Tsunematsu H. 1997. A new gene for blast resistance in rice cultivar, IR24. Rice Genet Newsl, 14: 60-61. |
| [17] | Imbe T, Tsunematsu H, Kato H, Khush G S. 1998. Genetic analysis of blast resistance in IR varieties and resistant breeding strategy. In: Tharreau D, Lebrun M H, Talbot N J, Notteghem J L. Advances in Rice Blast Research: Proceedings of the 2nd International Rice Blast Conference. Montpellier, France: 1-8. |
| [18] | International Standardization Organization. 2007. Rice: Determination of Amylose Content. Part 2: Routine Methods (ISO6647-2). International Standardization Organization. |
| [19] | IRRI (International Rice Research Institute). 2013. Statistical Tool for Agricultural Research (STAR): Plant Breeding, Genetics, and Biotechnology. Los Baños, the Philippines: IRRI. |
| [20] | IRRI (International Rice Research Institute). 2014. Standard Evaluation System for Rice (SES). 5th edn. Los Baños, the Philippines: IRRI: 57. |
| [21] | Jeung J U, Kim B R, Cho Y C, Han S S, Moon H P, Lee Y T, Jena K K. 2007. A novel gene Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice. Theor Appl Genet, 115: 1163-1177. |
| [22] | Kato T, Endo I, Yano M, Sasaki T, Inoue M, Kudo S. 2002. Mapping of quantitative trait loci for field resistance to rice blast in upland rice, ‘Sensho’. Breeding Res, 4(3): 119-124. |
| [23] | Kawasaki-Tanaka A, Fukuta Y. 2014. Genetic variation in resistance to blast disease (Pyricularia oryzae Cavara) in Japanese rice(Oryza sativa L.), as determined using a differential system. Breeding Sci, 64(2): 183-192. |
| [24] | Kiyosawa S, Ling Z. 2001. Genetic studies on rice blast relationships. In: Sreenivasaprasad S, Johnson R. Major Fungal Disease in Rice. Dordrecht: Kluwer Academic: 145-162. |
| [25] | Kou Y J, Wang S P. 2012. Toward an understanding of the molecular basis of quantitative disease resistance in rice. J Biotechnol, 159(4): 283-290. |
| [26] | Li Y C, Yuan L P. 1985. Genetic analysis of fertility restoration in male sterile lines of rice. In: Banta S J. Rice Genetics. Manila, the Philippines: International Rice Research Institute: 617-632. |
| [27] | Miura K, Agetsuma M, Kitano H, Yoshimura A, Matsuoka M, Jacobsen S E, Ashikari M. 2009. A metastable DWARF1 epigenetic mutant affecting plant stature in rice. Proc Natl Acad Sci USA, 106: 11218-11223. |
| [28] | Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins N C, Panstruga R, Schulze-Lefert P. 2002. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol, 129(3): 1076-1085. |
| [29] | Prahalada G D, Ramkumar G, Hechanova S L, Vinarao R, Jena K K. 2017. Exploring key blast and bacterial blight resistance genes in genetically diverse rice accessions through molecular and phenotypic evaluation. Crop Sci, 57: 1881-1892. |
| [30] | Ram S, Mishra B. 2010. Cereals: Processing and Nutritional Quality. New Delhi, India: New India Publishing Agency: 326. |
| [31] | Rick C M, Smith P G. 1953. Novel variation in tomato species hybrids. Am Nat, 87: 359-375. |
| [32] | Riesberg L H, Archer M A, Wayne R K. 1999. Transgressive segregation, adaptation and speciation. Heredity, 83: 363-372. |
| [33] | Sallaud C, Lorieux M, Roumen E, Tharreau D, Berruyer R, Svestasrani P, Garsmeur O, Ghesquiere A, Notteghem J L. 2003. Identification of five new blast resistance genes in the highly blast-resistant rice variety IR64 using a QTL mapping strategy. Theor Appl Genet, 106: 794-803. |
| [34] | Scheuermann K K, Raimondi J V, Marschalek R, Andrade A, Wickert E. 2012. Magnaporthe oryzae genetic diversity and its outcomes on the search for durable resistance. In: Caliskan M. The Molecular Basis of Plant Genetic Diversity. Intech Open Access Publisher: 331-356. |
| [35] | Selisana S M, Yanoria M J, Quime B, Chaipanya C, Lu G, Opulencia R, Wang G L, Mitchel T, Correll J, Talbot N J, Leung H, Zhou B. 2017. Avirulence (AVR) gene-based diagnosis complements existing pathogen surveillance tools for effective deployment of resistance (R) genes against rice blast disease. Phytopathology, 107(6): 711-720. |
| [36] | Séré Y, Onasanya A, Afolabi A, Mignouna H D, Akator K. 2007. Genetic diversity of the blast fungus Magnaporthe grisea(Hebert) Barr, in Burkina Faso. Afr J Biotechnol, 6: 2568-2577. |
| [37] | Sharma T R, Rai A K, Gupta S K, Vijayan J, Devanna B N, Ray S. 2012. Rice blast management through host-plant resistance: Retrospect and prospects. Agric Res, 1: 37-52. |
| [38] | Shim J, Torollo G, Angeles-Shim R B, Cabunagan R C, Choi I R, Yeo U S, Ha W G. 2015. Rice tungro spherical virus resistance into photoperiod-insensitive japonica rice by marker-assisted selection. Breeding Sci, 65(4): 345-351. |
| [39] | Suh J P, Roh J H, Cho Y C, Han S S, Kim Y G, Jena K K. 2009. The Pi40 gene for durable resistance to rice blast and molecular analysis of Pi40-advanced backcross breeding lines. Phytopathology, 99(3): 243-250. |
| [40] | Teleblanco-Yanoria M J, Imber T, Kato H, Tsunematsu H, Ebron L A, Vera Cruz C M, Kobayashi N, Fukuta Y. 2008. A set of standard differential blast isolates (Magnaporthe grisea (Hebert) Barr.) from the Philippines for rice (Oryza sativa L.) resistance. J Agric Res Quart, 42(1): 23-34. |
| [41] | Teleblanco-Yanoria M J, Koide Y, Fukuta Y, Imbe T, Tsunematsu H, Kato H, Ebron L A, Nguyen T M N, Kobayashi N. 2011. A set of differential lines of indica-type rice variety CO39 as differential varieties for blast resistance. Mol Breeding, 27(3): 357-373. |
| [42] | Uga Y, Okuno K, Yano M. 2011. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J Exp Bot, 62(8): 2485-2494. |
| [43] | Uga Y, Yamamoto E, Kanno N, Kawai S, Mizubayashi T, Fukuoka S. 2013. A major QTL controlling deep rooting on rice chromosome 4. Sci Rep, 3: 3040. |
| [44] | Vasudevan K, Vera Cruz C M, Gruissem W, Bhullar N K. 2014. Large scale germplasm screening for identification of novel blast resistance sources. Front Plant Sci, 5: 505. |
| [45] | Vega U, Frey K J. 1980. Transgressive segregation in inter- and intra-specific crosses of barley. Euphytica, 29(3): 585-594. |
| [46] | Wallwork H, Johnson R. 1984. Transgressive segregation for resistance to yellow rust in wheat. Euphytica, 33(1): 123-132. |
| [47] | Wang C L, Ulloa M, Mullens T R, Yu J Z, Roberts P A. 2012. QTL analysis for transgressive resistance to root-knot nematode in interspecific cotton (Gossypium spp) progeny derived from susceptible parents. PLoS One, 7: e344874. |
| [48] | Yasuda N, Mitsunaga T, Hayashi K, Koizumi S, Fujita Y. 2015. Effects of pyramiding quantitative resistance genes pi21, Pi34 and Pi35 on rice leaf blast disease. Plant Dis, 99(7): 904-909. |
| [49] | Yue B, Xue W Y, Xiong L Z, Yu X Q, Luo L J, Cui K H, Jin D M, Xing Y Z, Zhang Q F. 2006. Genetic basis of drought resistance at reproductive stage in rice: Separation of drought tolerance from drought avoidance. Genetics, 172(2): 1213-1228. |
| [50] | Zeigler R S, Tohme J, Nelson R, Levy M, Corre F J. 1994. Lineage exclusion: A proposal for linking blast population analysis to breeding. In: Zeigler R S, Leong S A, Teeng P S. Rice Blast Disease. Wallingford: CAB International: 267-292. |
| [51] | Zeng Y X, Xia L Z, Wen Z H, Ji Z J, Zeng D L, Qian Q, Yang C D. 2015. Mapping resistant QTLs for rice sheath blight disease with a doubled haploid population. J Integr Agric, 14(5): 801-810. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||