
Rice Science ›› 2024, Vol. 31 ›› Issue (3): 317-327.DOI: 10.1016/j.rsci.2024.02.001
收稿日期:2023-09-22
接受日期:2024-02-05
出版日期:2024-05-28
发布日期:2024-06-04
. [J]. Rice Science, 2024, 31(3): 317-327.
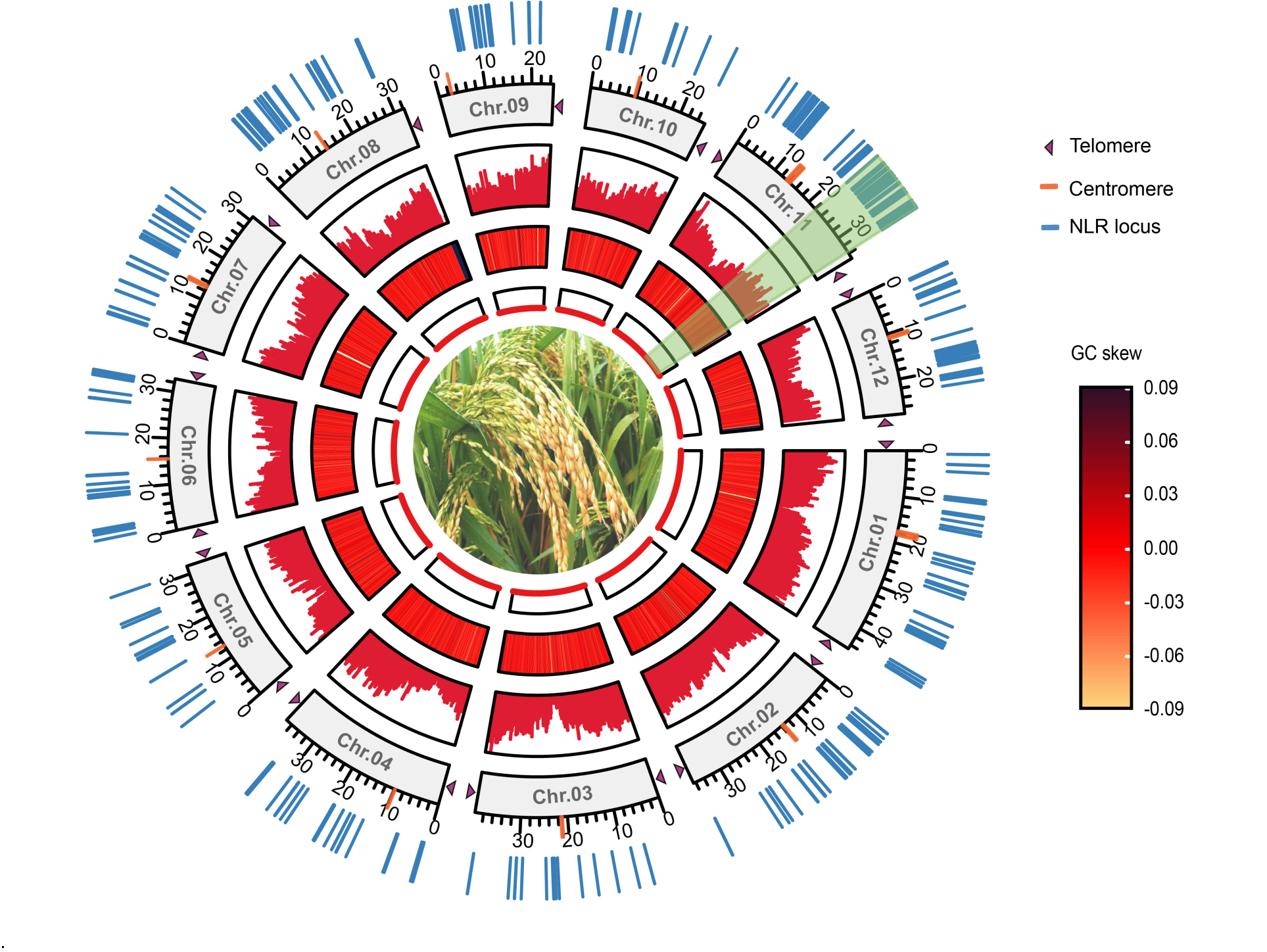
Fig. 1. Genomic landscape of rice Zhonghui 8015. From inside to outside: Distribution of the genome (1st), GC skew of the whole genome (2nd) (i.e. the offset of GC content in single-stranded DNA), gene density (3rd), chromosome length (4th, orange ticks represent predicted centromere regions, and purple triangles represent 5′- and 3′-telomeres), the light green region is a significant nucleotide-binding leucine-rich repeat (NLR) gene enriched region at the telomeric end of chromosome 11 (5th), and distribution of identified NLR genes in the genome (6th).
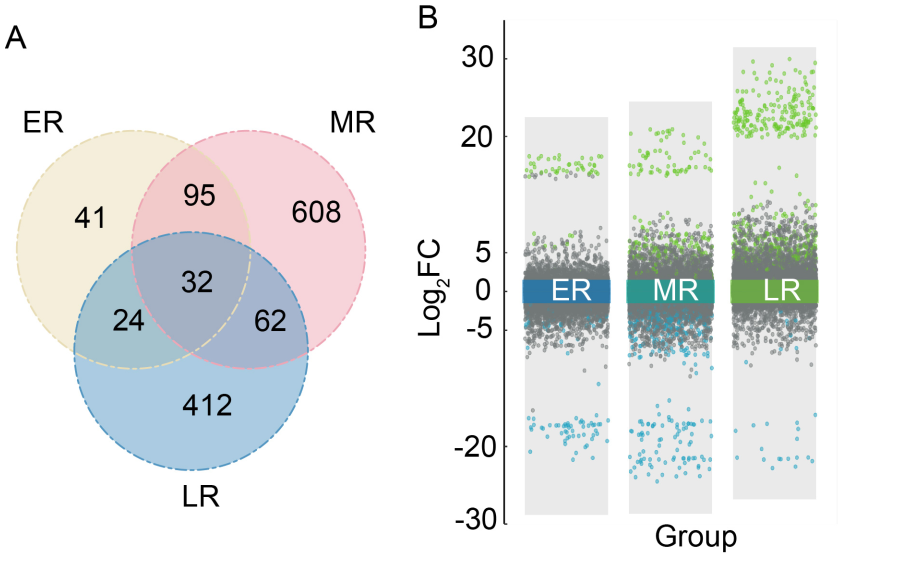
Fig. 2. Differentially expressed genes (DEGs) in brown planthopper (BPH) infestation groups compared with control group. A, Venn plot for number of DEGs. B, Volcano plot for DEGs. Each point represents a DEG. The green points represent genes with up-regulated expression compared with the control RNA-seq (with none BPH infestation), the blue points represent down-regulated genes, and the gray points are genes without significant difference in expression. ER, Early stage RNA-seq; MR, Middle stage RNA-seq; LR, Late stage RNA-seq; FC, Fold change.
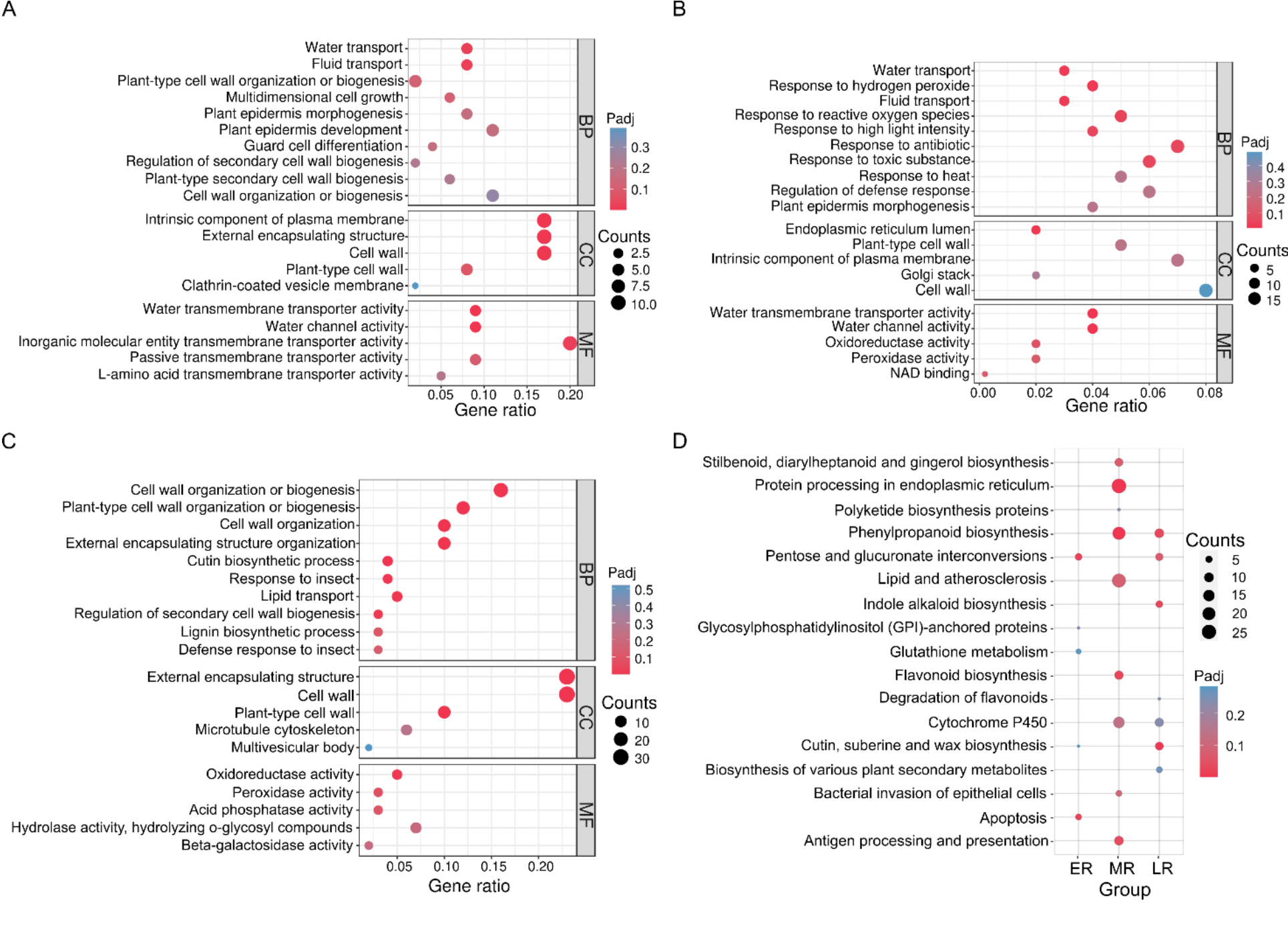
Fig. 3. Enrichment results of differentially expressed genes (DEGs) in three brown planthopper (BPH) infestation groups. A, Gene Ontology (GO) enrichment of ER vs CR. B, GO enrichment of MR vs CR. C, GO enrichment of LR vs CR. D, Kyoto Encyclopedia of Genes and Genomes enrichment results of three groups vs CR. CR, Control RNA-seq, with no BPH infestation; ER, Early stage RNA-seq, 12 h after BPH infestation; MR, Middle stage RNA-seq, 48 h after BPH infestation; LR, Late stage RNA-seq, 96 h after BPH infestation; BP, Biological process; CC, Cell component; MF, Molecular function. The size of the bubbles reflects the number of genes enriched in the KEGG pathway/GO term, while color reflects the significance of the enrichment.
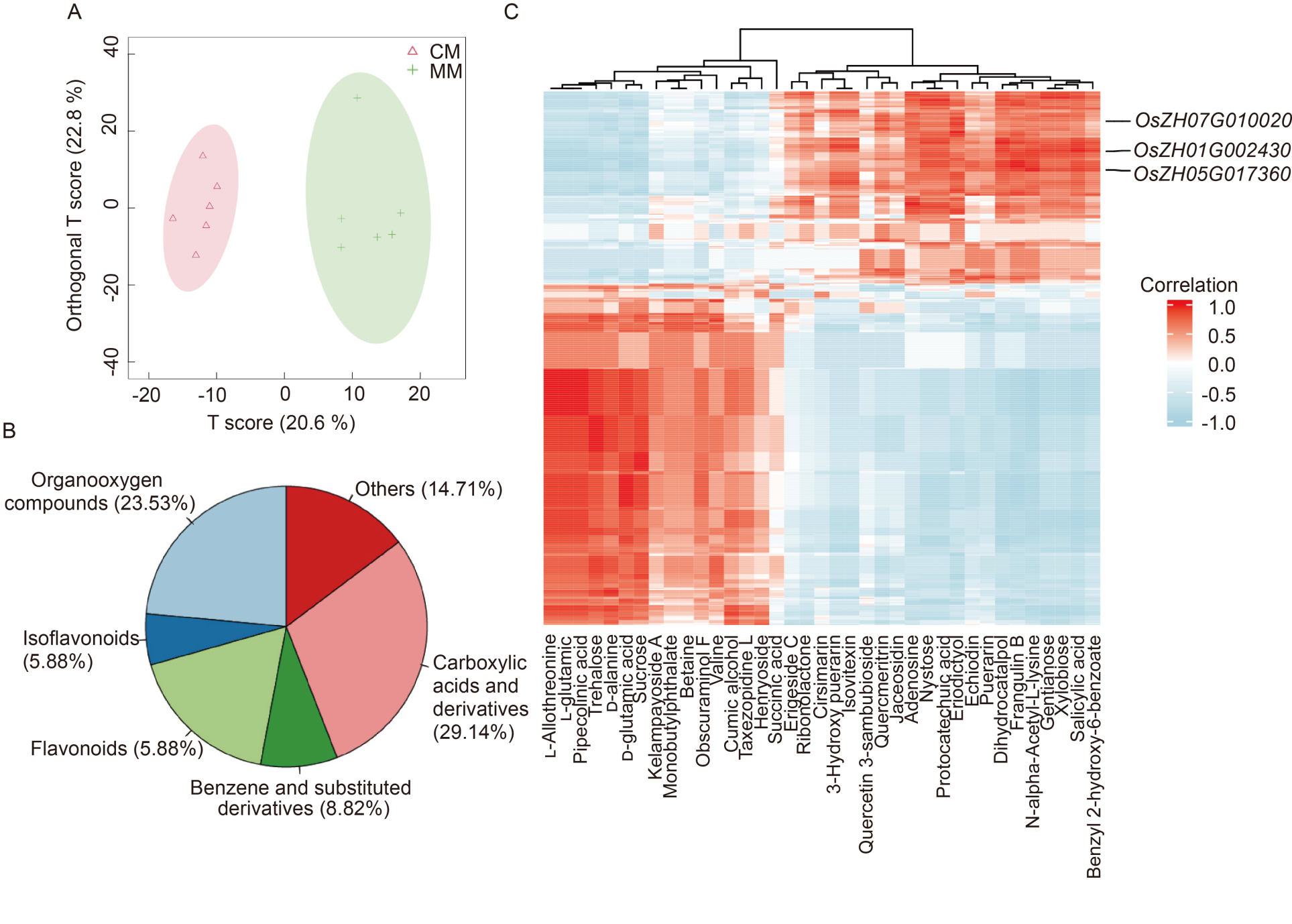
Fig. 4. Integrated analysis of metabolomics data (at the middle stage, 48 h brown planthopper infestation). A, Orthogonal Partial Least Squares Discrimination Analysis for discriminating between different brown planthopper infestation groups. CM, Control metabolism; MM, Middle stage metabolism, 48 h brown planthopper infestation. B, Categorization of differentially abundant metabolites. C, Heatmap of Spearman correlation coefficients between differentially abundant metabolite and differentially expressed gene levels. Each row represents a gene, and each column represents a metabolite.
| [1] | Benson G. 1999. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res, 27(2): 573-580. |
| [2] | Cheng H Y, Concepcion G T, Feng X W, Zhang H W, Li H. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods, 18(2): 170-175. |
| [3] | Cheng X Y, Zhu L L, He G C. 2013. Towards understanding of molecular interactions between rice and the brown planthopper. Mol Plant, 6(3): 621-634. |
| [4] | Cheng Z K, Dong F G, Langdon T, Ouyang S, Buell C R, Gu M H, Blattner F R, Jiang J M. 2002. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell, 14(8): 1691-1704. |
| [5] | Chern M, Canlas P E, Fitzgerald H A, Ronald P C. 2005. Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J, 43(5): 623-635. |
| [6] | Chern M, Bai W, Sze-To W H, Canlas P E, Bartley L E, Ronald P C. 2012. A rice transient assay system identifies a novel domain in NRR required for interaction with NH1/OsNPR1 and inhibition of NH1-mediated transcriptional activation. Plant Methods, 8(1): 6. |
| [7] | Copetti D, Zhang J W, El Baidouri M, Gao D Y, Wang J, Barghini E, Cossu R M, Angelova A, Roffler S, Ohyanagi H, Wicker T, Fan C Z, Zuccolo A, Chen M S, de Oliveira A C, Han B, Henry R, Hsing Y I, Kurata N, Wang W, Jackson S A, Panaud O, Wing R A. 2015. RiTE database: A resource database for genus-wide rice genomics and evolutionary biology. BMC Genomics, 16(1): 538. |
| [8] | Dai Z Y, Tan J, Zhou C, Yang X F, Yang F, Zhang S J, Sun S C, Miao X X, Shi Z Y. 2019. The OsmiR396-OsGRF8-OsF3H- flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol J, 17(8): 1657-1669. |
| [9] | Flynn J M, Hubley R, Goubert C, Rosen J, Clark A G, Feschotte C, Smit A F. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci USA, 117(17): 9451-9457. |
| [10] | Gao Z Y, Zhao S C, He W M, Guo L B, Peng Y L, Wang J J, Guo X S, Zhang X M, Rao Y C, Zhang C, Dong G J, Zheng F Y, Lu C X, Hu J, Zhou Q, Liu H J, Wu H Y, Xu J, Ni P X, Zeng D L, Liu D H, Tian P, Gong L H, Ye C, Zhang G H, Wang J, Tian F K, Xue D W, Liao Y, Zhu L, Chen M S, Li J Y, Cheng S H, Zhang G Y, Wang J, Qian Q. 2013. Dissecting yield-associated loci in super hybrid rice by resequencing recombinant inbred lines and improving parental genome sequences. Proc Natl Acad Sci USA, 110(35): 14492-14497. |
| [11] | Goff S A, Ricke D, Lan T H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange B M, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood T C, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller R M, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296(5565): 92-100. |
| [12] | Guo J P, Xu C X, Wu D, Zhao Y, Qiu Y F, Wang X X, Ouyang Y D, Cai B D, Liu X, Jing S L, Shangguan X X, Wang H Y, Ma Y H, Hu L, Wu Y, Shi S J, Wang W L, Zhu L L, Xu X, Chen R Z, Feng Y Q, Du B, He G C. 2018. Bph6 encodes an exocyst- localized protein and confers broad resistance to planthoppers in rice. Nat Genet, 50(2): 297-306. |
| [13] | Guo J P, Wang H Y, Guan W, Guo Q, Wang J, Yang J, Peng Y X, Shan J H, Gao M Y, Shi S J, Shangguan X X, Liu B F, Jing S L, Zhang J, Xu C X, Huang J, Rao W W, Zheng X H, Wu D, Zhou C, Du B, Chen R Z, Zhu L L, Zhu Y X, Walling L L, Zhang Q F, He G C. 2023. A tripartite rheostat controls self-regulated host plant resistance to insects. Nature, 618: 799-807. |
| [14] | Haas B J, Delcher A L, Mount S M, Wortman J R, Smith Jr R K, Hannick L I, Maiti R, Ronning C M, Rusch D B, Town C D, Salzberg S L, White O. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res, 31(19): 5654-5666. |
| [15] | Jing S L, Zhang L, Ma Y H, Liu B F, Zhao Y, Yu H J, Zhou X, Qin R, Zhu L L, He G C. 2014. Genome-wide mapping of virulence in brown planthopper identifies loci that break down host plant resistance. PLoS One, 9(6): e98911. |
| [16] | Kawahara Y, de la Bastide M, Hamilton J P, Kanamori H, McCombie W R, Ouyang S, Schwartz D C, Tanaka T, Wu J Z, Zhou S G, Childs K L, Davidson R M, Lin H N, Quesada- Ocampo L, Vaillancourt B, Sakai H, Lee S S, Kim J, Numa H, Itoh T, Buell C R, Matsumoto T. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice, 6(1): 4. |
| [17] | Kim D, Paggi J M, Park C, Bennett C, Salzberg S L. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol, 37(8): 907-915. |
| [18] | Kourelis J, Sakai T, Adachi H, Kamoun S. 2021. RefPlantNLR is a comprehensive collection of experimentally validated plant disease resistance proteins from the NLR family. PLoS Biol, 19(10): e3001124. |
| [19] | Li X, Kapos P, Zhang Y L. 2015. NLRs in plants. Curr Opin Immunol, 32: 114-121. |
| [20] | Lin Y Z, Ye C, Li X Z, Chen Q Y, Wu Y, Zhang F, Pan R, Zhang S J, Chen S X, Wang X, Cao S, Wang Y Z, Yue Y, Liu Y S, Yue J Y. 2023. quarTeT: A telomere-to-telomere toolkit for gap-free genome assembly and centromeric repeat identification. Hortic Res, 10(8): uhad127. |
| [21] | Liu M Y, Hong G J, Li H J, Bing X L, Chen Y M, Jing X F, Gershenzon J, Lou Y G, Baldwin I T, Li R. 2023. Sakuranetin protects rice from brown planthopper attack by depleting its beneficial endosymbionts. Proc Natl Acad Sci USA, 120(23): e2305007120. |
| [22] | Love M I, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15(12): 550. |
| [23] | Murata K, Kitano T, Yoshimoto R, Takata R, Ube N, Ueno K, Ueno M, Yabuta Y, Teraishi M, Holland C K, Jander G, Okumoto Y, Mori N, Ishihara A. 2020. Natural variation in the expression and catalytic activity of a naringenin 7-O-methyltransferase influences antifungal defenses in diverse rice cultivars. Plant J, 101(5): 1103-1117. |
| [24] | Ou S J, Chen J F, Jiang N. 2018. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res, 46(21): e126. |
| [25] | Qin P, Lu H W, Du H L, Wang H, Chen W L, Chen Z, He Q, Ou S J, Zhang H Y, Li X Z, Li X X, Li Y, Liao Y, Gao Q, Tu B, Yuan H, Ma B T, Wang Y P, Qian Y W, Fan S J, Li W T, Wang J, He M, Yin J J, Li T, Jiang N, Chen X W, Liang C Z, Li S G. 2021. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell, 184(13): 3542-3558. |
| [26] | Quinlan A R. 2014. BEDTools: The swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics, 47: 11.12. 1-11.12.34. |
| [27] | Rao S S P, Huntley M H, Durand N C, Stamenova E K, Bochkov I D, Robinson J T, Sanborn A L, Machol I, Omer A D, Lander E S, Aiden E L. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 159(7): 1665-1680. |
| [28] | Rhie A, Walenz B P, Koren S, Phillippy A M. 2020. Merqury: Reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol, 21(1): 245. |
| [29] | Schmittgen T D, Livak K J. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc, 3: 1101-1108. |
| [30] | Shang L G, Li X X, He H Y, Yuan Q L, Song Y N, Wei Z R, Lin H, Hu M, Zhao F L, Zhang C, Li Y H, Gao H S, Wang T Y, Liu X P, Zhang H, Zhang Y, Cao S M, Yu X M, Zhang B T, Zhang Y, Tan Y Q, Qin M, Ai C, Yang Y X, Zhang B, Hu Z Q, Wang H R, Lv Y, Wang Y X, Ma J, Wang Q, Lu H W, Wu Z, Liu S L, Sun Z Y, Zhang H L, Guo L B, Li Z C, Zhou Y F, Li J Y, Zhu Z F, Xiong G S, Ruan J, Qian Q. 2022. A super pan-genomic landscape of rice. Cell Res, 32(10): 878-896. |
| [31] | Shang L G, He W C, Wang T Y, Yang Y X, Xu Q, Zhao X J, Yang L B, Zhang H, Li X X, Lv Y, Chen W, Cao S, Wang X M, Zhang B, Liu X P, Yu X M, He H Y, Wei H, Leng Y, Shi C L, Guo M L, Zhang Z P, Zhang B T, Yuan Q L, Qian H G, Cao X L, Cui Y, Zhang Q Q, Dai X F, Liu C C, Guo L B, Zhou Y F, Zheng X M, Ruan J, Cheng Z K, Pan W H, Qian Q. 2023. A complete assembly of the rice Nipponbare reference genome. Mol Plant, 16(8): 1232-1236. |
| [32] | Shi S J, Wang H Y, Nie L Y, Tan D, Zhou C, Zhang Q, Li Y, Du B, Guo J P, Huang J, Wu D, Zheng X H, Guan W, Shan J H, Zhu L L, Chen R Z, Xue L J, Walling L L, He G C. 2021. Bph30 confers resistance to brown planthopper by fortifying sclerenchyma in rice leaf sheaths. Mol Plant, 14(10): 1714-1732. |
| [33] | Shumate A, Wong B, Pertea G, Pertea M. 2022. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS Comput Biol, 18(6): e1009730. |
| [34] | Simão F A, Waterhouse R M, Ioannidis P, Kriventseva E V, Zdobnov E M. 2015. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31(19): 3210-3212. |
| [35] | Song J M, Xie W Z, Wang S, Guo Y X, Koo D H, Kudrna D, Gong C B, Huang Y C, Feng J W, Zhang W H, Zhou Y, Zuccolo A, Long E, Lee S, Talag J, Zhou R, Zhu X T, Yuan D J, Udall J, Xie W B, Wing R A, Zhang Q F, Poland J, Zhang J W, Chen L L. 2021. Two gap-free reference genomes and a global view of the centromere architecture in rice. Mol Plant, 14(10): 1757-1767. |
| [36] | Sudheeran P K, Ovadia R, Galsarker O, Maoz I, Sela N, Maurer D, Feygenberg O, Oren Shamir M, Alkan N. 2020. Glycosylated flavonoids: Fruit’s concealed antifungal arsenal. New Phytol, 225(4): 1788-1798. |
| [37] | Tan Q Y, Zhu H T, Liu H, Ni Y R, Wu S Z, Luan X, Liu J W, Yang W F, Yang Z F, Zeng R Z, Liu G F, Wang S K, Zhang G Q. 2022. Fine mapping of QTLs for stigma exsertion rate from Oryza glaberrima by chromosome segment substitution. Rice Sci, 29(1): 55-66. |
| [38] | Tarailo-Graovac M, Chen N S. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinform, 25: 4.10. 1-4.10.14. |
| [39] | Wan H L, Liu H B, Zhang J Y, Lyu Y, Li Z R, He Y Z, Zhang X L, Deng X X, Brotman Y, Fernie A R, Cheng Y J, Wen W W. 2020. Lipidomic and transcriptomic analysis reveals reallocation of carbon flux from cuticular wax into plastid membrane lipids in a glossy “Newhall” navel orange mutant. Hortic Res, 7: 41. |
| [40] | Wang S, Lei C L, Wang J L, Ma J, Tang S, Wang C L, Zhao K J, Tian P, Zhang H, Qi C Y, Cheng Z J, Zhang X, Guo X P, Liu L L, Wu C Y, Wan J M. 2017. SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J Exp Bot, 68(5): 899-913. |
| [41] | Wang S, Gao S H, Nie J Y, Tan X Y, Xie J H, Bi X C, Sun Y, Luo S N, Zhu Q H, Geng J N, Liu W F, Lin Q, Cui P, Hu S N, Wu S Y. 2022. Improved 93-11 genome and time-course transcriptome expand resources for rice genomics. Front Plant Sci, 12: 769700. |
| [42] | Xu M Y, Guo L D, Gu S Q, Wang O, Zhang R, Peters B A, Fan G Y, Liu X, Xu X, Deng L, Zhang Y W. 2020. TGS-GapCloser: A fast and accurate gap closer for large genomes with low coverage of error-prone long reads. GigaScience, 9(9): giaa094. |
| [43] | Yang Q Q, Zhang Y X, Xue P, Wen X X, Liu L, Xu P, Zhan X D, Cao L Y, Cheng S H, Wu W X. 2022. QTL mapping for plant height using introgression lines derived from Zhonghui 8015 and wild rice (Oryza rufipogon). Rice Sci, 29(6): 503-506. |
| [44] | Yırtıcı Ü, Ergene A, Atalar M N, Adem Ş. 2022. Phytochemical composition, antioxidant, enzyme inhibition, antimicrobial effects, and molecular docking studies of Centaurea sivasica. S Afr N J Bot, 144: 58-71. |
| [45] | Yu G C, Wang L G, Han Y Y, He Q Y. 2012. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS, 16(5): 284-287. |
| [46] | Yu J, Hu S N, Wang J, Wong G K S, Li S G, Liu B, Deng Y J, Dai L, Zhou Y, Zhang X Q, Cao M L, Liu J, Sun J D, Tang J B, Chen Y J, Huang X B, Lin W, Ye C, Tong W, Cong L J, Geng J N, Han Y J, Li L, Li W, Hu G Q, Huang X G, Li W J, Li J, Liu Z W, Li L, Liu J P, Qi Q H, Liu J S, Li L, Li T, Wang X G, Lu H, Wu T T, Zhu M, Ni P X, Han H, Dong W, Ren X Y, Feng X L, Cui P, Li X R, Wang H, Xu X, Zhai W X, Xu Z, Zhang J S, He S J, Zhang J G, Xu J C, Zhang K L, Zheng X W, Dong J H, Zeng W Y, Tao L, Ye J, Tan J, Ren X D, Chen X W, He J, Liu D F, Tian W, Tian C G, Xia H A, Bao Q Y, Li G, Gao H, Cao T, Wang J, Zhao W M, Li P, Chen W, Wang X D, Zhang Y, Hu J F, Wang J, Liu S, Yang J, Zhang G Y, Xiong Y Q, Li Z J, Mao L, Zhou C S, Zhu Z, Chen R S, Hao B L, Zheng W M, Chen S Y, Guo W, Li G J, Liu S Q, Tao M, Wang J, Zhu L H, Yuan L P, Yang H M. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science, 296(5565): 79-92. |
| [47] | Yu Z C, Chen Y M, Zhou Y, Zhang Y L, Li M Y, Ouyang Y D, Chebotarov D, Mauleon R, Zhao H, Xie W B, McNally K L, Wing R A, Guo W L, Zhang J W. 2023. Rice Gene Index: A comprehensive pan-genome database for comparative and functional genomics of Asian rice. Mol Plant, 16(5): 798-801. |
| [48] | Zdobnov E M, Apweiler R. 2001. InterProScan: An integration platform for the signature-recognition methods in InterPro. Bioinformatics, 17(9): 847-848. |
| [49] | Zhan X D, Yu P, Cao L Y, Shen X H, Chen D B, Cheng S H. 2015. Breeding of excellent high-yield and high-quality hybrid rice new combination Nei5you 8015. Bull Agric Sci Techn, (1): 102-103. (in Chinese) |
| [50] | Zhang C, Shi C N, Chen D, Wu J G. 2018. Rice ragged stunt virus propagation and infection on rice plants. Bio-protocol, 8(20): e3060. |
| [51] | Zhang F, Xue H Z, Dong X R, Li M, Zheng X M, Li Z K, Xu J L, Wang W S, Wei C C. 2022. Long-read sequencing of 111 rice genomes reveals significantly larger pan-genomes. Genome Res, 32(5): 853-863. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||