
Rice Science ›› 2021, Vol. 28 ›› Issue (4): 391-401.DOI: 10.1016/j.rsci.2021.05.009
• Research Papers • Previous Articles Next Articles
Guangchen Zhang1,#, Zimeng Liu1,#, Youhong Liu2, Noriyuki Kuya3, Yuchen Hua1, Hongru Shi4, Weilin Zhao1, Yuqing Han1, Toshio Yamamoto5, Wenfu Chen1( ), Jian Sun1(
), Jian Sun1( )
)
Received:2020-06-15
Accepted:2020-11-10
Online:2021-07-28
Published:2021-07-28
About author:#These authors contributed equally to this work
Guangchen Zhang, Zimeng Liu, Youhong Liu, Noriyuki Kuya, Yuchen Hua, Hongru Shi, Weilin Zhao, Yuqing Han, Toshio Yamamoto, Wenfu Chen, Jian Sun. iTRAQ-Based Proteomics Investigation of Critical Response Proteins in Embryo and Coleoptile During Rice Anaerobic Germination[J]. Rice Science, 2021, 28(4): 391-401.
Add to citation manager EndNote|Ris|BibTeX
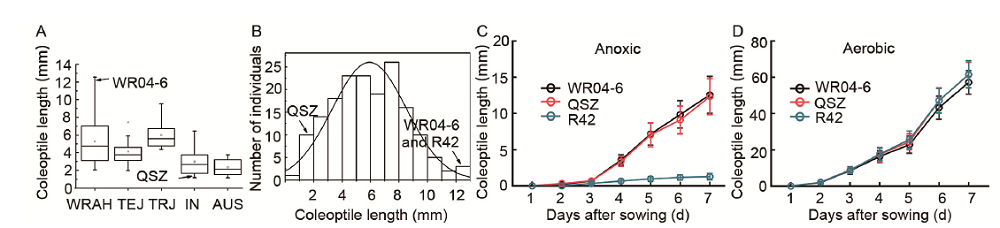
Fig. 1. Phenotypic identification of anaerobic germination. A, Distribution of coleoptile length in five subgroups under anoxic conditions on 7 d after sowing. WRAH, Weedy rice at Asian high latitudes; TEJ, temperature japonica; TRJ, tropical japonica; IN, indica; AUS, aus; QSZ, Qishanzhan. B, Frequency distribution of coleoptile length of QSZ, R42, WR04-6 and recombinant inbred lines under anoxic conditions on 7 d after sowing. C, Dynamic of coleoptile elongation of QSZ, R42 and WR04-6 under anoxic conditions. D, Dynamic of coleoptile elongation of QSZ, R42 and WR04-6 under aerobic conditions.
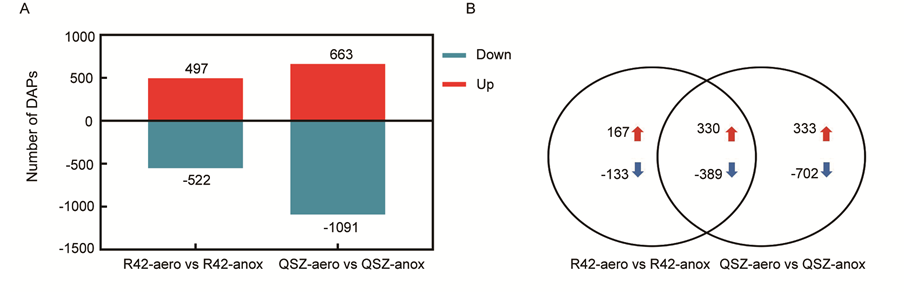
Fig. S2. Statistics of differentially abundant proteins in two comparison groups. A, Number of up-regulated and down-regulated DAPs in two comparison groups. B, Venn diagram of DAPs in R42 and QSZ groups. The overlapping region indicates the number of common response DAPs and the region in the R42 comparison group alone indicate tolerance-specific DAPs.DAPs, Differentially abundant proteins.
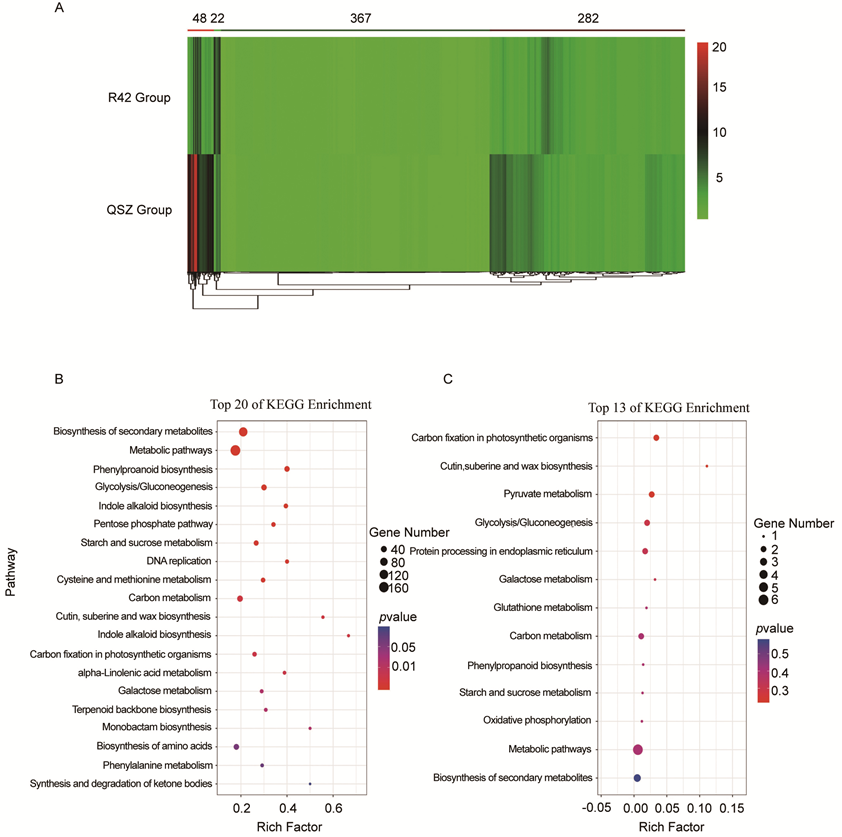
Fig. S3. Analysis of 719 common response differentially abundant proteins. A, Hierarchical cluster analysis of 719 common response DAPs. B, KEGG enrichment analysis for DAPs with concordant expression patterns with 282 up-regulated and 367 down-regulated. C, KEGG enrichment analysis for DAPs with different expression patterns with 48 up-regulated and 22 down-regulated.
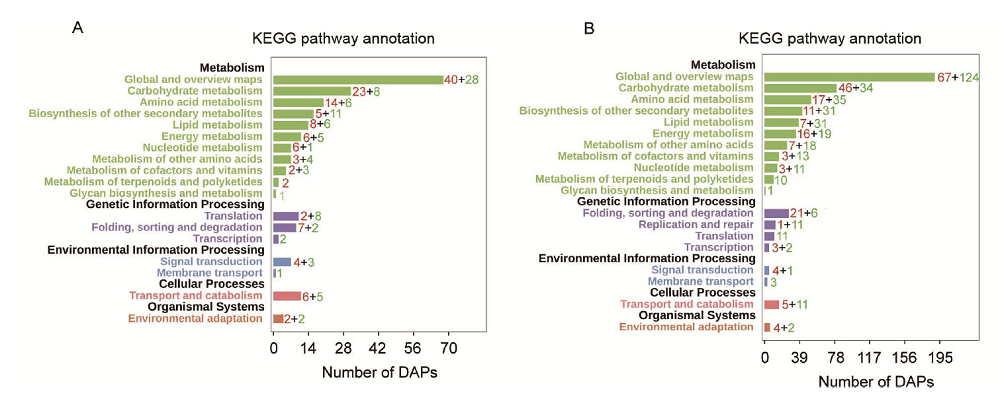
Fig. 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differentially abundant proteins during rice anaerobic germination. A, KEGG analysis of tolerance-specific differentially abundant proteins (DAPs). B, KEGG analysis of common response DAPs.
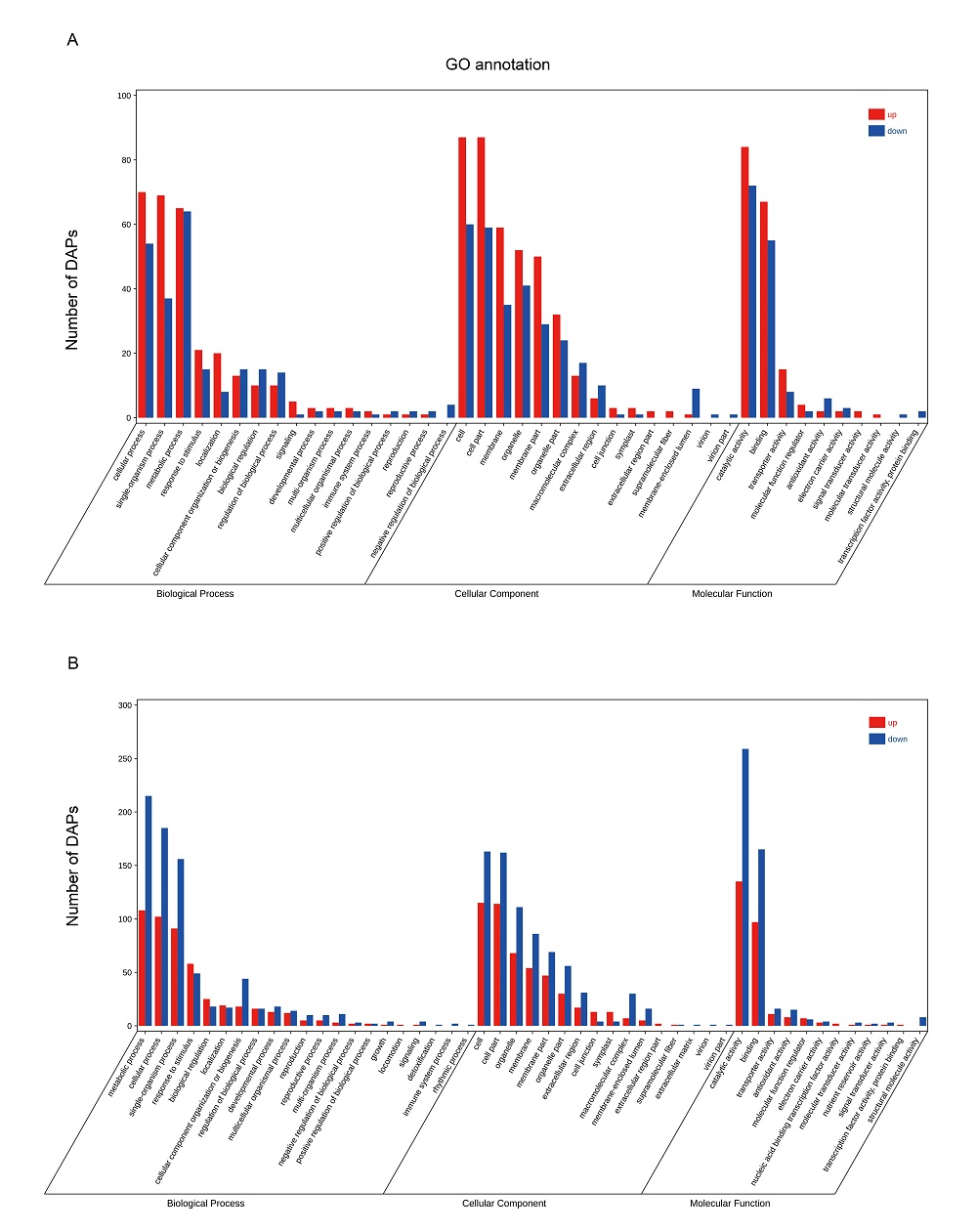
Fig. S4. Gene ontology (GO) analysis of differentially abundant proteins during rice anaerobic germination. A, GO analysis of tolerance-specific DAPs. B, GO analysis of common response DAPs.
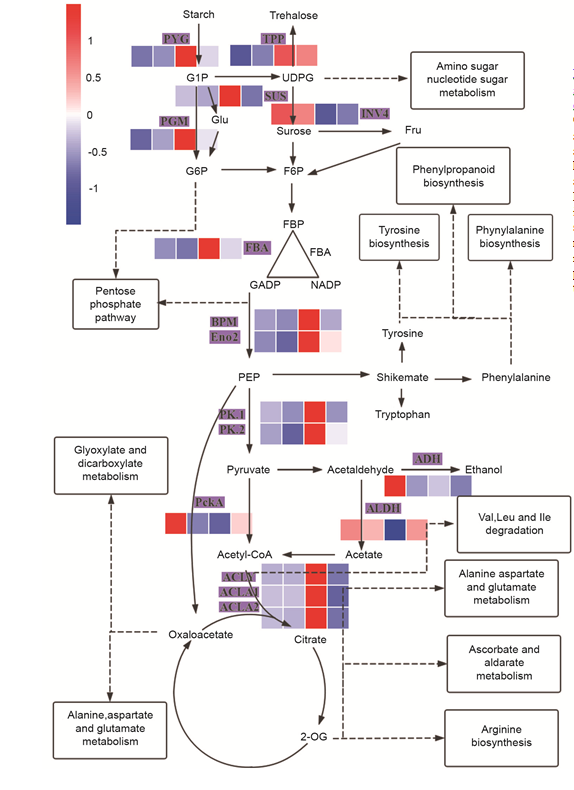
Fig. S5. Changes of differentially abundant proteins (DAPs) in carbohydrate metabolism. Characters with purple backgrounds are DAPs. The four colored squares around the DAP names ordered from left to right indicate expression abundance of R42-aero, QSZ-aero, R42-anox and QSZ-anox. Arrows indicate the direction of metabolites, solid lines represent metabolites to metabolites, and dotted lines represent metabolites to KEGG pathways that were enriched in tolerance-specific DAPs.
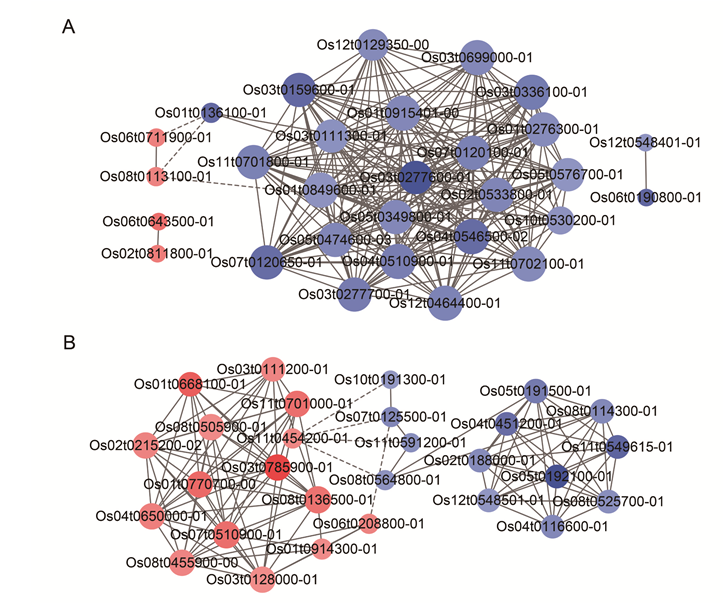
Fig. S6. rotein-protein interaction network. A, The protein-protein interaction network of the top 30 common response DAPs with the highest FC value. B, The protein-protein interaction network of the top 30 tolerance specific DAPs with the highest FC value. Red and blue nodes represent relatively up-regulated and down-regulated DAPs, and the color depth of each node represents the fold change: a dark color means a higher number of Log2 fold change, a light color means a lower number of Log2 fold change. The size of each node indicates the number of interacting proteins. Solid lines represent positive correlations and dotted lines represent negative correlations.
| Number | Protein ID | Regulated | IP | Description | ||
|---|---|---|---|---|---|---|
| 1 | Os03t0277600-01 | Down | 19 | Similar to (clone wusl1032) mRNA sequence. | ||
| 2 | Os06t0190800-01 | Down | 1 | Similar to Glutamate receptor. | ||
| 3 | Os01t0136100-01 | Down | 4 | 16.9 kDa class I heat shock protein 1. | ||
| 4 | Os03t0159600-01 | Down | 20 | Similar to Rab28 protein. | ||
| 5 | Os04t0546500-02 | Down | 21 | Similar to Oleosin. | ||
| 6 | Os07t0120650-01 | Down | 20 | Protein of unknown function DUF538 family protein. | ||
| 7 | Os03t0336100-01 | Down | 21 | 11-S plant seed storage protein family protein. | ||
| 8 | Os03t0277700-01 | Down | 20 | Protein of unknown function DUF26 domain containing protein. | ||
| 9 | Os06t0643500-01 | Up | 1 | Similar to ADR11 protein (Fragment). | ||
| 10 | Os04t0510900-01 | Down | 21 | Similar to Embryo-specific protein 1 (ATS1). | ||
| 11 | Os11t0701800-01 | Down | 20 | Chitinase (EC 3.2.1.14) III C10701-rice (EC 3.2.1.14) (Class III chitinase homologue (OsChib3H-a) H-). | ||
| 12 | Os07t0120100-01 | Down | 20 | Protein of unknown function DUF538 family protein. | ||
| 13 | Os12t0464400-01 | Down | 21 | Similar to Oxidoreductase, short chain dehydrogenase/reductase family protein, expressed. | ||
| 14 | Os02t0533800-01 | Down | 20 | Similar to ATPase inhibitor. | ||
| 15 | Os05t0474600-03 | Down | 20 | Similar to Aldose reductase-related protein (EC 1.1.1.21). | ||
| 16 | Os05t0349800-01 | Down | 20 | Embryonic abundant protein 1. | ||
| 17 | Os03t0699000-01 | Down | 21 | Oleosin 18 kDa (OSE721). | ||
| 18 | Os01t0915401-00 | Down | 21 | Proteinase inhibitor I25, cystatin domain containing protein. | ||
| 19 | Os12t0129350-00 | Down | 17 | Similar to Arabinoxylan arabinofuranohydrolase isoenzyme AXAH-I. | ||
| 20 | Os11t0702100-01 | Down | 21 | Similar to Class III chitinase homologue (OsChib3H-h) (Fragment). | ||
| 21 | Os01t0276300-01 | Down | 20 | Similar to Group 3 late embryogenesis abundant protein (Fragment). | ||
| 22 | Os02t0811800-01 | Up | 1 | Similar to Cinnamoyl-CoA reductase (EC 1.2.1.44). | ||
| 23 | Os06t0711900-01 | Up | 2 | Bifunctional inhibitor/plant lipid transfer protein/seed storage domain containing protein. | ||
| 24 | Os03t0111300-01 | Down | 19 | Nonspecific lipid-transfer protein 2 (nsLTP2) (7 kDa lipid transfer protein). | ||
| 25 | Os08t0509400-01 | Down | 0 | Similar to Amygdalin hydrolase isoform AH I precursor (EC 3.2.1.117). | ||
| 26 | Os10t0530200-01 | Down | 13 | Similar to Glutathione S-transferase TSI-1 (EC 2.5.1.18) (Glutathione S- transferase 1). | ||
| 27 | Os01t0849600-01 | Down | 21 | Similar to ENOD18 protein (Fragment). | ||
| 28 | Os12t0548401-01 | Down | 1 | Similar to Proteinase inhibitor. | ||
| 29 | Os08t0113100-01 | Up | 3 | Similar to Fructokinase (Fragment). | ||
| 30 | Os05t0576700-01 | Down | 19 | Oleosin family protein. | ||
Table S3. Top 30 common response DAPs
| Number | Protein ID | Regulated | IP | Description | ||
|---|---|---|---|---|---|---|
| 1 | Os03t0277600-01 | Down | 19 | Similar to (clone wusl1032) mRNA sequence. | ||
| 2 | Os06t0190800-01 | Down | 1 | Similar to Glutamate receptor. | ||
| 3 | Os01t0136100-01 | Down | 4 | 16.9 kDa class I heat shock protein 1. | ||
| 4 | Os03t0159600-01 | Down | 20 | Similar to Rab28 protein. | ||
| 5 | Os04t0546500-02 | Down | 21 | Similar to Oleosin. | ||
| 6 | Os07t0120650-01 | Down | 20 | Protein of unknown function DUF538 family protein. | ||
| 7 | Os03t0336100-01 | Down | 21 | 11-S plant seed storage protein family protein. | ||
| 8 | Os03t0277700-01 | Down | 20 | Protein of unknown function DUF26 domain containing protein. | ||
| 9 | Os06t0643500-01 | Up | 1 | Similar to ADR11 protein (Fragment). | ||
| 10 | Os04t0510900-01 | Down | 21 | Similar to Embryo-specific protein 1 (ATS1). | ||
| 11 | Os11t0701800-01 | Down | 20 | Chitinase (EC 3.2.1.14) III C10701-rice (EC 3.2.1.14) (Class III chitinase homologue (OsChib3H-a) H-). | ||
| 12 | Os07t0120100-01 | Down | 20 | Protein of unknown function DUF538 family protein. | ||
| 13 | Os12t0464400-01 | Down | 21 | Similar to Oxidoreductase, short chain dehydrogenase/reductase family protein, expressed. | ||
| 14 | Os02t0533800-01 | Down | 20 | Similar to ATPase inhibitor. | ||
| 15 | Os05t0474600-03 | Down | 20 | Similar to Aldose reductase-related protein (EC 1.1.1.21). | ||
| 16 | Os05t0349800-01 | Down | 20 | Embryonic abundant protein 1. | ||
| 17 | Os03t0699000-01 | Down | 21 | Oleosin 18 kDa (OSE721). | ||
| 18 | Os01t0915401-00 | Down | 21 | Proteinase inhibitor I25, cystatin domain containing protein. | ||
| 19 | Os12t0129350-00 | Down | 17 | Similar to Arabinoxylan arabinofuranohydrolase isoenzyme AXAH-I. | ||
| 20 | Os11t0702100-01 | Down | 21 | Similar to Class III chitinase homologue (OsChib3H-h) (Fragment). | ||
| 21 | Os01t0276300-01 | Down | 20 | Similar to Group 3 late embryogenesis abundant protein (Fragment). | ||
| 22 | Os02t0811800-01 | Up | 1 | Similar to Cinnamoyl-CoA reductase (EC 1.2.1.44). | ||
| 23 | Os06t0711900-01 | Up | 2 | Bifunctional inhibitor/plant lipid transfer protein/seed storage domain containing protein. | ||
| 24 | Os03t0111300-01 | Down | 19 | Nonspecific lipid-transfer protein 2 (nsLTP2) (7 kDa lipid transfer protein). | ||
| 25 | Os08t0509400-01 | Down | 0 | Similar to Amygdalin hydrolase isoform AH I precursor (EC 3.2.1.117). | ||
| 26 | Os10t0530200-01 | Down | 13 | Similar to Glutathione S-transferase TSI-1 (EC 2.5.1.18) (Glutathione S- transferase 1). | ||
| 27 | Os01t0849600-01 | Down | 21 | Similar to ENOD18 protein (Fragment). | ||
| 28 | Os12t0548401-01 | Down | 1 | Similar to Proteinase inhibitor. | ||
| 29 | Os08t0113100-01 | Up | 3 | Similar to Fructokinase (Fragment). | ||
| 30 | Os05t0576700-01 | Down | 19 | Oleosin family protein. | ||
| Number | Protein ID | Regulated | IP | Description |
|---|---|---|---|---|
| 1 | Os05t0192100-01 | Down | 8 | Similar to Stem 28 kDa glycoprotein. |
| 2 | Os05t0399400-00 | Down | 0 | Chitinase 9. |
| 3 | Os03t0785900-01 | Up | 11 | Similar to Glutathione-S-transferase. |
| 4 | Os11t0549615-01 | Down | 8 | Metallophosphoesterase domain containing protein. |
| 5 | Os04t0451200-01 | Down | 8 | UDP-glucuronosyl/UDP-glucosyltransferase family protein. |
| 6 | Os01t0668100-01 | Up | 9 | Similar to Arabinogalactan-like protein. |
| 7 | Os01t0770700-00 | Up | 11 | Similar to Copper transporter 1. |
| 8 | Os05t0191500-01 | Down | 9 | Similar to stem 28 kDa glycoprotein. |
| 9 | Os07t0510900-01 | Up | 12 | Similar to L-ascorbate oxidase. |
| 10 | Os11t0701000-01 | Up | 11 | Class III chitinase homologue (OsChib3H-c). |
| 11 | Os08t0136500-01 | Up | 12 | Protein of unknown function DUF26 domain containing protein. |
| 12 | Os07t0125500-01 | Down | 4 | Allergen V5/Tpx-1 related family protein. |
| 13 | Os04t0116600-01 | Down | 8 | Glucose/ribitol dehydrogenase family protein. |
| 14 | Os08t0114300-01 | Down | 9 | D-arabinono-1 |
| 15 | Os02t0188000-01 | Down | 8 | UDP-glucuronosyl/UDP-glucosyltransferase family protein. |
| 16 | Os11t0591200-01 | Down | 3 | Similar to 3-ketoacyl-CoA synthase. |
| 17 | Os08t0525700-01 | Down | 8 | Conserved hypothetical protein. |
| 18 | Os04t0650000-01 | Up | 11 | Similar to Oryzain alpha chain precursor (EC 3.4.22.-). |
| 19 | Os08t0564800-01 | Down | 4 | Coactivator CBP |
| 20 | Os02t0215200-02 | Up | 11 | Similar to Histone deacetylase. |
| 21 | Os06t0208800-01 | Up | 4 | Lysin motif-containing protein |
| 22 | Os08t0502400-01 | Up | 0 | FAS1 domain domain containing protein. |
| 23 | Os08t0455900-00 | Up | 11 | Putative copper transporter 5.2. |
| 24 | Os03t0111200-01 | Up | 9 | Similar to Remorin. |
| 25 | Os12t0548501-01 | Down | 8 | Similar to CI2C. |
| 26 | Os10t0191300-01 | Down | 2 | Similar to PR-1a pathogenesis related protein (Hv-1a) precursor. |
| 27 | Os11t0454200-01 | Up | 4 | Dehydrin RAB 16B. |
| 28 | Os08t0505900-01 | Up | 11 | Leucine-rich repeat (LRR) protein |
| 29 | Os01t0914300-01 | Up | 5 | Plant lipid transfer protein/seed storage/trypsin-alpha amylase inhibitor domain containing protein. |
| 30 | Os03t0128000-01 | Up | 11 | FAS1 domain containing protein. |
Table S4. Top 30 tolerance-specific DAPs
| Number | Protein ID | Regulated | IP | Description |
|---|---|---|---|---|
| 1 | Os05t0192100-01 | Down | 8 | Similar to Stem 28 kDa glycoprotein. |
| 2 | Os05t0399400-00 | Down | 0 | Chitinase 9. |
| 3 | Os03t0785900-01 | Up | 11 | Similar to Glutathione-S-transferase. |
| 4 | Os11t0549615-01 | Down | 8 | Metallophosphoesterase domain containing protein. |
| 5 | Os04t0451200-01 | Down | 8 | UDP-glucuronosyl/UDP-glucosyltransferase family protein. |
| 6 | Os01t0668100-01 | Up | 9 | Similar to Arabinogalactan-like protein. |
| 7 | Os01t0770700-00 | Up | 11 | Similar to Copper transporter 1. |
| 8 | Os05t0191500-01 | Down | 9 | Similar to stem 28 kDa glycoprotein. |
| 9 | Os07t0510900-01 | Up | 12 | Similar to L-ascorbate oxidase. |
| 10 | Os11t0701000-01 | Up | 11 | Class III chitinase homologue (OsChib3H-c). |
| 11 | Os08t0136500-01 | Up | 12 | Protein of unknown function DUF26 domain containing protein. |
| 12 | Os07t0125500-01 | Down | 4 | Allergen V5/Tpx-1 related family protein. |
| 13 | Os04t0116600-01 | Down | 8 | Glucose/ribitol dehydrogenase family protein. |
| 14 | Os08t0114300-01 | Down | 9 | D-arabinono-1 |
| 15 | Os02t0188000-01 | Down | 8 | UDP-glucuronosyl/UDP-glucosyltransferase family protein. |
| 16 | Os11t0591200-01 | Down | 3 | Similar to 3-ketoacyl-CoA synthase. |
| 17 | Os08t0525700-01 | Down | 8 | Conserved hypothetical protein. |
| 18 | Os04t0650000-01 | Up | 11 | Similar to Oryzain alpha chain precursor (EC 3.4.22.-). |
| 19 | Os08t0564800-01 | Down | 4 | Coactivator CBP |
| 20 | Os02t0215200-02 | Up | 11 | Similar to Histone deacetylase. |
| 21 | Os06t0208800-01 | Up | 4 | Lysin motif-containing protein |
| 22 | Os08t0502400-01 | Up | 0 | FAS1 domain domain containing protein. |
| 23 | Os08t0455900-00 | Up | 11 | Putative copper transporter 5.2. |
| 24 | Os03t0111200-01 | Up | 9 | Similar to Remorin. |
| 25 | Os12t0548501-01 | Down | 8 | Similar to CI2C. |
| 26 | Os10t0191300-01 | Down | 2 | Similar to PR-1a pathogenesis related protein (Hv-1a) precursor. |
| 27 | Os11t0454200-01 | Up | 4 | Dehydrin RAB 16B. |
| 28 | Os08t0505900-01 | Up | 11 | Leucine-rich repeat (LRR) protein |
| 29 | Os01t0914300-01 | Up | 5 | Plant lipid transfer protein/seed storage/trypsin-alpha amylase inhibitor domain containing protein. |
| 30 | Os03t0128000-01 | Up | 11 | FAS1 domain containing protein. |
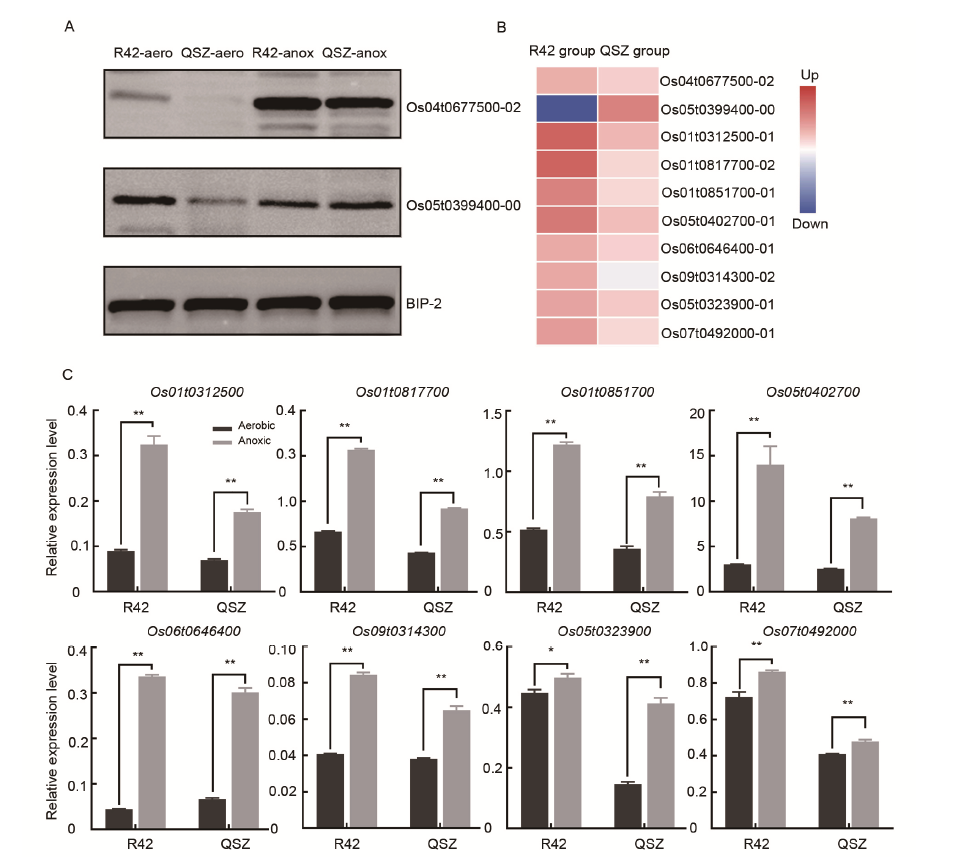
Fig. 3. Validation of isobaric tags for relative and absolute quantitation (iTRAQ) results. A, Western blot analysis of pyruvate kinase (Os04t0677500-02) and chitinase 9 (Os05t0399400-00) for R42 under aerobic condition (R42-aero), Qishanzhan under aerobic condition (QSZ-aero), R42 under anoxic condition (R42-anox) and Qishanzhan under anoxic condition (QSZ-anox). Accumulated level of BIP-2 was used as a loading control. B, Heat map showing the fold changes of selected differentially abundant proteins abundance in aerobic and anoxic conditions. The fold change was obtained relative to the aerobic conditions. C, Quantitative real-time PCR analysis of the transcription levels of eight differentially abundant protein related genes. Data are Mean ± SD (n = 3). *, P ≤ 0.05; **, P ≤ 0.01.
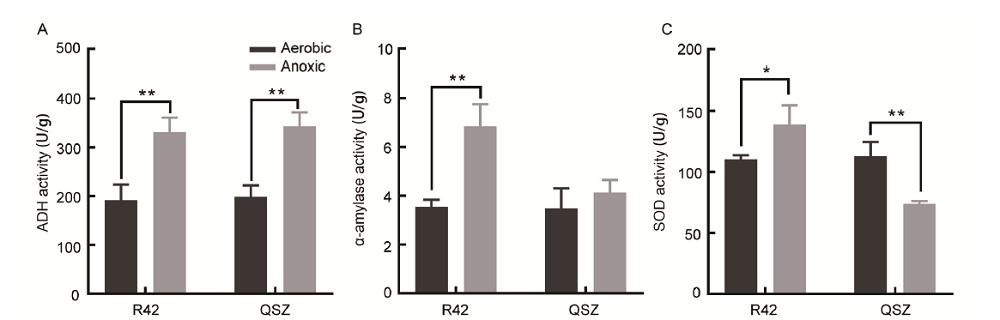
Fig. 4. Activity comparison of enzymes between R42 and Qishanzhan (QSZ) under aerobic and anoxic conditions. A, Alcohol dehydrogenase (ADH) activity. B, α-amylase activity. C, Superoxide dismutase (SOD) activity. Data are Mean ± SD (n = 3). *, P ≤ 0.05; **, P ≤ 0.01.
| ID | Forward | Reverse |
|---|---|---|
| Os01t0312500 | GTGCTGACAAGTGAAGAAAGAG | CATTTCGGCCCAAAAACAAAAG |
| Os01t0817700 | ACGGTGGTATCCAGATTCTTAC | CCATGGAAGTTCATTACGGTTG |
| Os01t0851700 | CTTTTGGCACCTATGACTATGC | CGTATTTAGGAGACAGGGCTAG |
| Os05t0402700 | GAGGTCATCGCTGAGTACAC | CTTGTTCATGGCGTTGAGGTTC |
| Os06t0646400 | TCTCAAGCTCTCATCAACACAT | ATTAGCCATATAAGTCGGCACA |
| Os09t0314300 | TTCTGACACCTTTAGCAAAAGC | CATACACATTGGCATTCTCAGG |
| Os05t0323900 | CTACGTCGCCAACTACAACAAG | AAGATCGAATGATTGACATGGC |
| Os07t0492000 | CTCTGGTTCCCTGAAGGTTTAG | AAGAGGTGATCGATTATAGGCG |
| OsACT | GAACTGGTATGGTCAAGGCTG | ACACGGAGCTCGTTGTAGAAG |
Table S5. Primers used in this study.
| ID | Forward | Reverse |
|---|---|---|
| Os01t0312500 | GTGCTGACAAGTGAAGAAAGAG | CATTTCGGCCCAAAAACAAAAG |
| Os01t0817700 | ACGGTGGTATCCAGATTCTTAC | CCATGGAAGTTCATTACGGTTG |
| Os01t0851700 | CTTTTGGCACCTATGACTATGC | CGTATTTAGGAGACAGGGCTAG |
| Os05t0402700 | GAGGTCATCGCTGAGTACAC | CTTGTTCATGGCGTTGAGGTTC |
| Os06t0646400 | TCTCAAGCTCTCATCAACACAT | ATTAGCCATATAAGTCGGCACA |
| Os09t0314300 | TTCTGACACCTTTAGCAAAAGC | CATACACATTGGCATTCTCAGG |
| Os05t0323900 | CTACGTCGCCAACTACAACAAG | AAGATCGAATGATTGACATGGC |
| Os07t0492000 | CTCTGGTTCCCTGAAGGTTTAG | AAGAGGTGATCGATTATAGGCG |
| OsACT | GAACTGGTATGGTCAAGGCTG | ACACGGAGCTCGTTGTAGAAG |
| [1] | Angaji S A, Septiningsih E M, Mackill D J, Ismail A M. 2010. QTLs associated with tolerance of flooding during germination in rice (Oryza sativa L.). Euphytica, 172(2): 159-168. |
| [2] | Bailey-Serres J, Voesenek L A C J. 2008. Flooding stress: Acclimations and genetic diversity. Annu Rev Plant Biol, 59: 313-339. |
| [3] | Baltazar M D, Ignacio J C I, Thomson M J, Ismail A M, Mendioro M S, Septiningsih E M. 2014. QTL mapping for tolerance of anaerobic germination from IR64 and the aus landrace Nanhi using SNP genotyping. Euphytica, 197(2): 251-260. |
| [4] | Cosgrove D J. 2005. Growth of the plant cell wall. Nat Rev Mol Cell Biol, 6(11): 850-861. |
| [5] | Gara L D, de Pinto M C, Arrigoni O. 1997. Ascorbate synthesis and ascorbate peroxidase activity during the early stage of wheat germination. Physiol Plant, 100(4): 894-900. |
| [6] | Hamoud Y A, Wang Z C, Guo X P, Shaghaleh H, Sheteiwy M, Chen S, Qiu R J, Elbashier M M A. 2019. Effect of irrigation regimes and soil texture on the potassium utilization efficiency of rice. Agronomy, 9(2): 100. |
| [7] | He D L, Yang P F. 2013. Proteomics of rice seed germination. Front Plant Sci, 4: 246. |
| [8] | Ho V T, Tran A N, Cardarelli F, Perata P, Pucciariello C. 2017. A calcineurin B-like protein participates in low oxygen signalling in rice. Funct Plant Biol, 44(9): 917-928. |
| [9] | Howell K A, Cheng K, Murcha M W, Jenkin L E, Millar A H, Whelan J. 2007. Oxygen initiation of respiration and mitochondrial biogenesis in rice. J Biol Chem, 282(21): 15619-15631. |
| [10] | Hsu S K, Tung C W. 2015. Genetic mapping of anaerobic germination- associated QTLs controlling coleoptile elongation in rice. Rice, 8: 38. |
| [11] | Hsu S K, Tung C W. 2017. RNA-seq analysis of diverse rice genotypes to identify the genes controlling coleoptile growth during submerged germination. Front Plant Sci, 8: 762. |
| [12] | Hu Q J, Lin C, Guan Y J, Sheteiwy M S, Hu W M, Hu J. 2017. Inhibitory effect of eugenol on seed germination and pre-harvest sprouting of hybrid rice (Oryza sativa L.). Sci Rep, 7(1): 5295. |
| [13] | Huang S B, Taylor N L, Narsai R, Eubel H, Whelan J, Millar A H. 2009. Experimental analysis of the rice mitochondrial proteome, its biogenesis, and heterogeneity. Plant Physiol, 149(2): 719-734. |
| [14] | Kawai M, Uchimiya H. 2000. Coleoptile senescence in rice (Oryza sativa L.). Ann Bot, 86(2): 405-414. |
| [15] | Komatsu S, Tougou M, Nanjo Y. 2015. Proteomic techniques and management of flooding tolerance in soybean. J Proteome Res, 14(9): 3768-3778. |
| [16] | Kretzschmar T, Pelayo M A F, Trijatmiko K R, Gabunada L F M, Alam R, Jimenez R, Mendioro M S, Slamet-Loedin I H, Sreenivasulu N, Bailey-Serres J, Ismail A M, Mackill D J, Septiningsih E M. 2015. A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat Plants, 1: 15124. |
| [17] | Kuya N, Sun J, Iijima K, Venuprasad R, Yamamoto T. 2019. Novel method for evaluation of anaerobic germination in rice and its application to diverse genetic collections. Breeding Sci, 69(4): 633-639. |
| [18] | Lasanthi-Kudahettige R, Magneschi L, Loreti E, Gonzali S, Licausi F, Novi G, Beretta O, Vitulli F, Alpi A, Perata P. 2007. Transcript profiling of the anoxic rice coleoptile. Plant Physiol, 144(1): 218-231. |
| [19] | Lee K W, Chen P W, Lu C A, Chen S, Ho T H D, Yu S M. 2009. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal, 2: ra61. |
| [20] | Liu J, Hasanuzzaman M, Sun H Z, Zhang J, Peng T, Sun H W, Xin Z Y, Zhao Q Z. 2020. Comparative morphological and transcriptomic responses of lowland and upland rice to root-zone hypoxia. Environ Exp Bot, 169: 103916. |
| [21] | Lu C A, Lin C C, Lee K W, Chen J L, Huang L F, Ho S L, Liu H J, Hsing Y I, Yu S M. 2007. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell, 19(8): 2484-2499. |
| [22] | Magneschi L, Perata P. 2009. Rice germination and seedling growth in the absence of oxygen. Ann Bot, 103(2): 181-196. |
| [23] | Manangkil O E, Vu H T T, Mori N, Yoshida S, Nakamura C. 2013. Mapping of quantitative trait loci controlling seedling vigor in rice (Oryza sativa L.) under submergence. Euphytica, 192(1): 63-75. |
| [24] | Miro B, Ismail A M. 2013. Tolerance of anaerobic conditions caused by flooding during germination and early growth in rice (Oryza sativa L.). Front Plant Sci, 4: 269. |
| [25] | Miro B, Longkumer T, Entila F D, Kohli A, Ismail A M. 2017. Rice seed germination underwater: Morpho-physiological responses and the bases of differential expression of alcoholic fermentation enzymes. Front Plant Sci, 8: 1857. |
| [26] | Mittler R. 2006. Abiotic stress, the field environment and stress combination. Trends Plant Sci, 11(1): 15-19. |
| [27] | Narsai R, Howell K A, Carroll A, Ivanova A, Millar A H, Whelan J. 2009. Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol, 151(1): 306-322. |
| [28] | Narsai R, Edwards J M, Roberts T H, Whelan J, Joss G H, Atwell B J. 2015. Mechanisms of growth and patterns of gene expression in oxygen-deprived rice coleoptiles. Plant J, 82(1): 25-40. |
| [29] | Nghi K N, Tondelli A, Valè G, Tagliani A, Marè C, Perata P, Pucciariello C. 2019. Dissection of coleoptile elongation in japonica rice under submergence through integrated genome-wide association mapping and transcriptional analyses. Plant Cell Environ, 42(6): 1832-1846. |
| [30] | Pandey A, Mann M. 2000. Proteomics to study genes and genomes. Nature, 405: 837-846. |
| [31] | Pearce D M E, Jackson M B. 1991. Comparison of growth responses of barnyard grass (Echinochloa oryzoides) and rice (Oryza sativa) to submergence, ethylene, carbon dioxide and oxygen shortage. Ann Bot, 68(3): 201-209. |
| [32] | Pujana M A, Han J D J, Starita L M, Stevens K N, Tewari M, Ahn J S, Rennert G, Moreno V, Kirchhoff T, Gold B, Assmann V, Elshamy W M, Rual J F, Levine D, Rozek L S, Gelman R S, Gunsalus K C, Greenbreg R A, Sobhian B, Bertin N, Venkatesan K, Ayivi- Guedehoussou N, Sole X, Hernandez P, Lazaro C, Nathanson K L, Weber B L, Cusick M E, Hill D E, Offit K, Livingston D M, Gruber S B, Parvin J D, Vidal M. 2007. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet, 39(11): 1338-1149. |
| [33] | Sedbrook J C, Kronebusch P J, Borisy G G, Trewavas A J, Masson P H. 1996. Transgenic AEQUORIN reveals organ-specific cytosolic Ca2+ responses to anoxia in Arabidopsis thaliana seedlings. Plant Physiol, 111(1): 243-257. |
| [34] | Septiningsih E M, Ignacio J C I, Sendon P M D, Sanchez D L, Ismail A M, Mackill D J. 2013. QTL mapping and confirmation for tolerance of anaerobic conditions during germination derived from the rice landrace Ma-Zhan Red. Theor Appl Genet, 126(5): 1357-1366. |
| [35] | Setter T L, Waters I. 2003. Review of prospects for germplasm improvement for waterlogging tolerance in wheat, barley and oats. Plant Soil, 253(1): 1-34. |
| [36] | Sheteiwy M S, Gong D T, Gao Y, Pan R H, Hu J, Guan Y J. 2018. Priming with methyl jasmonate alleviates polyethylene glycol- induced osmotic stress in rice seeds by regulating the seed metabolic profile. Environ Exp Bot, 153: 236‒248. |
| [37] | Sheteiwy M S, Shao H B, Qi W C, Hamoud Y A, Shaghaleh H, Khan N U, Yang R P, Tang B P. 2019. GABA-alleviated oxidative injury induced by salinity, osmotic stress and their combination by regulating cellular and molecular signals in rice. Int J Mol Sci, 20(22): 5709. |
| [38] | Shingaki-Wells R N, Huang S B, Taylor N L, Carroll A J, Zhou W X, Millar A H. 2011. Differential molecular responses of rice and wheat coleoptiles to anoxia reveal novel metabolic adaptations in amino acid metabolism for tissue tolerance. Plant Physiol, 156(4): 1706-1724. |
| [39] | Sun J, Qian Q, Ma D R, Xu Z J, Liu D, Du H B, Chen W F. 2013. Introgression and selection shaping the genome and adaptive loci of weedy rice in northern China. New Phytol, 197(1): 290-299. |
| [40] | Sun J, Ma D R, Tang L, Zhao M H, Zhang G C, Wang W J, Song J Y, Li X, Liu Z M, Zhang W X, Xu Q, Zhou Y C, Wu J Z, Yamamoto T, Dai F, Lei Y, Li S, Zhou G, Zheng H K, Xu Z J, Chen W F. 2019. Population genomic analysis and de novo assembly reveal the origin of weedy rice as an evolutionary game. Mol Plant, 12(5): 632-647. |
| [41] | Takahashi H, Saika H, Matsumura H, Nagamura Y, Tsutsumi N, Nishizawa N K, Nakazono M. 2011. Cell division and cell elongation in the coleoptile of rice alcohol dehydrogenase 1-deficient mutant are reduced under complete submergence. Ann Bot, 108(2): 253-261. |
| [42] | Vijayan J, Senapati S, Ray S, Chakraborty K, Molla K A, Basak N, Pradhan B, Yeasmin L, Chattopadhyay K, Sarkar R K. 2018. Transcriptomic and physiological studies identify cues for germination stage oxygen deficiency tolerance in rice. Environ Exp Bot, 147: 234-248. |
| [43] | Wiśniewski J R, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods, 6(5): 359-362. |
| [44] | Xiao G Y, Xiao Y L, Li J J, Deng L H, Weng L S, Meng Q C, Yu J H. 2019. High efficiency is a dominant target for current rice breeding. Chin J Rice Sci, 33(4): 287-292. (in Chinese with English abstract) |
| [45] | Yang J, Sun K, Li D X, Luo L X, Liu Y Z, Huang M, Yang G L, Liu H, Wang H, Chen Z Q, Guo T. 2019. Identification of stable QTLs and candidate genes involved in anaerobic germination tolerance in rice via high-density genetic mapping and RNA-Seq. BMC Genomics, 20(1): 355. |
| [46] | Ye N H, Wang F Z, Shi L, Chen M X, Cao Y Y, Zhu F Y, Wu Y Z, Xie L J, Liu T Y, Su Z Z, Xiao S, Zhang H, Yang J C, Gu H Y, Hou X X, Hu Q J, Yi H J, Zhu C X, Zhang J H, Liu Y G. 2018. Natural variation in the promoter of rice calcineurin B-like protein10 (OsCBL10) affects flooding tolerance during seed germination among rice subspecies. Plant J, 94(4): 612-625. |
| [47] | Yin X J, Sakata K, Komatsu S. 2014. Phosphoproteomics reveals the effect of ethylene in soybean root under flooding stress. J Proteome Res, 13(12): 5618-5634. |
| [48] | Yin X J, Komatsu S. 2015. Quantitative proteomics of nuclear phosphoproteins in the root tip of soybean during the initial stages of flooding stress. J Proteomics, 119: 183‒195. |
| [49] | Yu S M, Lee H T, Lo S F, Ho T H D. 2019. How does rice cope with too little oxygen during its early life? New Phytol, 229(1): 36-41. |
| [50] | Zhang M C, Lu Q, Wu W, Niu X J, Wang C H, Feng Y, Xu Q, Wang S, Yuan X P, Yu H Y, Wang Y P, Wei X H. 2017. Association mapping reveals novel genetic loci contributing to flooding tolerance during germination in indica rice. Front Plant Sci, 8: 678. |
| [1] | Mondal Satyen, Jamil Hasan M., Ahmed Tofayel, Giashuddin Miah M., C. Sta Cruz Pompe, M. Ismail Abdel. Effects of AG1 and AG2 QTLs on Nonstructural Carbohydrate and Seed Management Options for Rice Seedling Growth and Establishment under Flooding Stress [J]. Rice Science, 2020, 27(6): 515-528. |
| [2] | Deyong Zeng, Jie Cui, Yishu Yin, Meng Zhang, Shan Shan, Xin Gao, Yingchun Zhang, Yeqing Sun, Weihong Lu. Effects of Space Flight on Expression of Key Proteins in Rice Leaves [J]. Rice Science, 2020, 27(5): 423-433. |
| [3] | Mao-bai Li, Hui Wang, Li-ming Cao. Evaluation of Population Structure, Genetic Diversity and Origin of Northeast Asia Weedy Rice Based on Simple Sequence Repeat Markers [J]. Rice Science, 2015, 22(4): 180-188. |
| [4] | LI Xiao-yan, QIANG Sheng, SONG Xiao-ling, CAI Kun, SUN Yi-na, SHI Zhi-hua, DAI Wei-min. Allele Types of Rc Gene of Weedy Rice from Jiangsu Province, China [J]. RICE SCIENCE, 2014, 21(5): 252-261. |
| [5] | LU Yong-liang, Nilda R. BURGOS, WANG Wei-xia, YU Liu-qing. Transgene Flow from Glufosinate-Resistant Rice to Improved and Weedy Rice in China [J]. RICE SCIENCE, 2014, 21(5): 271-281. |
| [6] | ZHANG Juan, Nilda R. BURGOS, MA Kun, ZHOU Yong-jun, GENG Rui-mei, YU Liu-qing. Genetic Diversity and Relationship of Weedy Rice in Taizhou City, Jiangsu Province, China [J]. RICE SCIENCE, 2008, 15(4): 295-302 . |
| [7] | Mohammad Safdar BALOCH,*, Inayat Ullah AWAN, Gul HASSAN, Abdul Aziz KHAKWANI. Effect of Establishment Methods and Weed Management Practices on Some Growth Attributes of Rice [J]. RICE SCIENCE, 2006, 13(2): 131-140 . |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||