
Rice Science ›› 2021, Vol. 28 ›› Issue (5): 466-478.DOI: 10.1016/j.rsci.2021.07.007
• Research Paper • Previous Articles Next Articles
Yong Yang1,#, Qiujun Lin2,#, Xinyu Chen3, Weifang Liang4, Yuwen Fu2, Zhengjin Xu2, Yuanhua Wu2, Xuming Wang1, Jie Zhou1, Chulang Yu5, Chengqi Yan6, Qiong Mei2( ), Jianping Chen1,2,5(
), Jianping Chen1,2,5( )
)
Received:2020-08-19
Accepted:2021-03-02
Online:2021-09-28
Published:2021-09-28
About author:#These authors contributed equally to this work
Yong Yang, Qiujun Lin, Xinyu Chen, Weifang Liang, Yuwen Fu, Zhengjin Xu, Yuanhua Wu, Xuming Wang, Jie Zhou, Chulang Yu, Chengqi Yan, Qiong Mei, Jianping Chen. Characterization and Proteomic Analysis of Novel Rice Lesion Mimic Mutant with Enhanced Disease Resistance[J]. Rice Science, 2021, 28(5): 466-478.
Add to citation manager EndNote|Ris|BibTeX
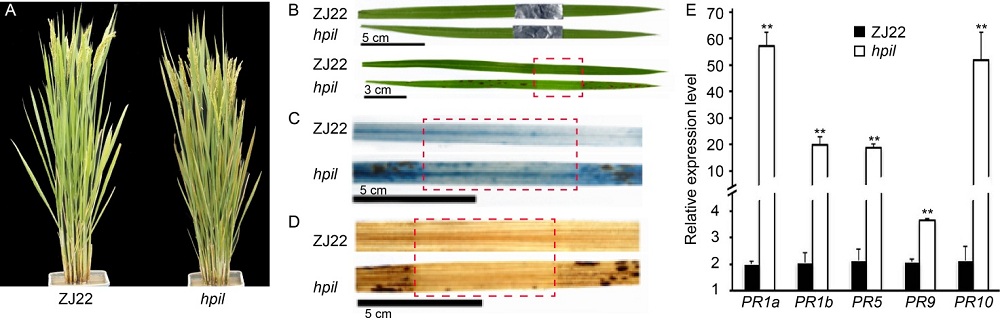
Fig. 1. Characterization of hpil and relative expression of pathogenesis-related proteins (PRs). A, Phenotypes of Zhejing 22 (ZJ22) and hpil. B, Leaves of ZJ22 and hpil. Shaded region is indicated by red box. C, Trypan blue staining of leaves of ZJ22 and hpil. Shaded region is indicated by red box. D, Diaminobenzidine staining of leaves of ZJ22 and hpil. Shaded region is indicated by red box.E, Relative expression of PRs in ZJ22 and hpil. Data are Mean ± SE (n = 3). **, P < 0.01.
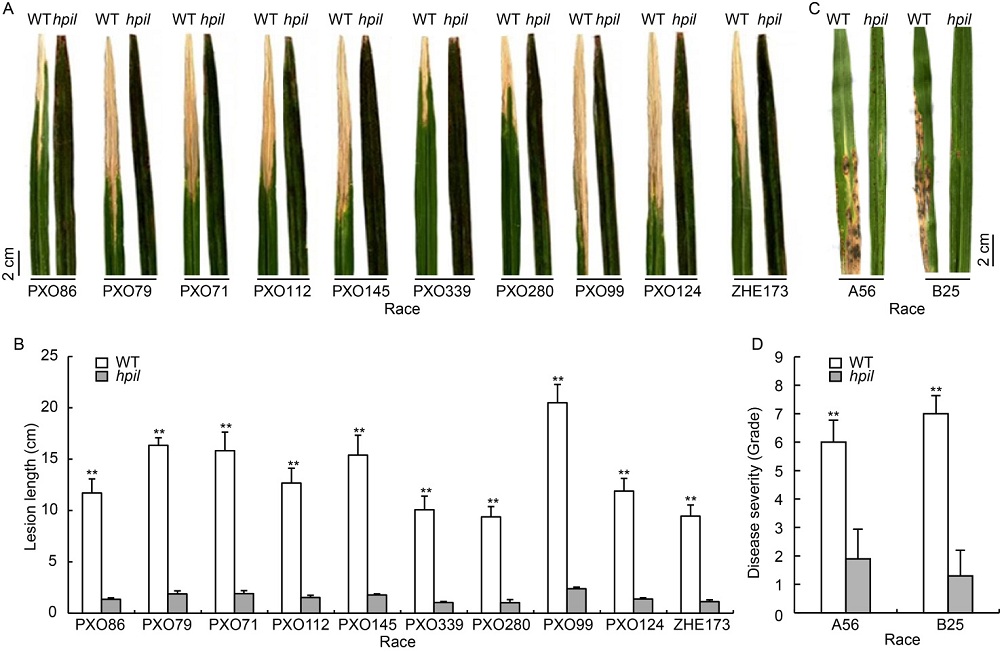
Fig. 2. High resistance of hpil to different races of Xoo and M. oryzae.A, Leaves of Zhejing 22 (WT) and hpil inoculated with 10 different races of Xoo. B, Lesion lengths caused by Xoo infection. Data are Mean ± SD (n = 6); **, P < 0.01.C, Leaves of WT and hpil inoculated with two different races of M. oryzae. D, Disease severity of leaves inoculated with M. oryzae. Data are Mean ± SD (n = 10); **, P < 0.01.
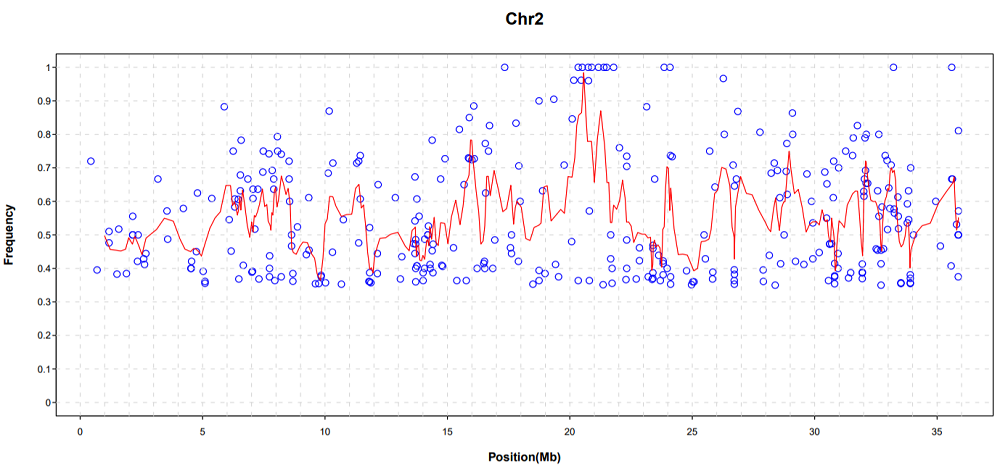
Fig. S1. Genotype frequency profile of progeny on chromosome 2.The x-coordinate represents the coordinates of the chromosomes. The ordinate represents the frequency of the mutant genotype. The red line represents the corresponding frequency values of the sliding window in different regions of the chromosome.

Fig. S2. Numbers of differentially expressed protein spots on hpil in comparison with wild-type plants.Protein spots (≥ 1.5-fold, P < 0.05) detected in leaves from three positions (1, 2 and 3).
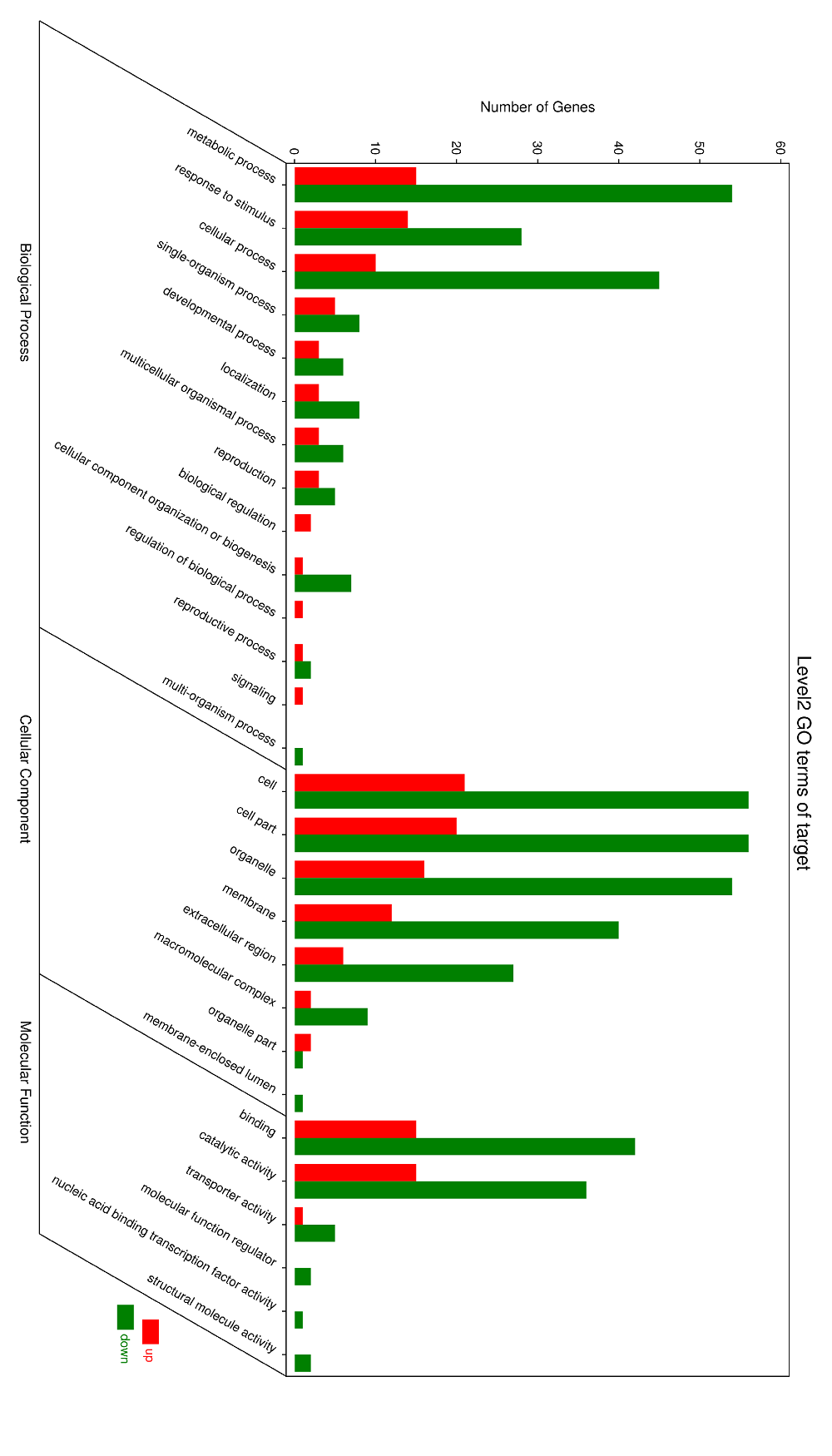
Fig. S3. Gene ontology classification of differently expressed proteins.Red bar represent up-regulated spots, green bar represent down-regulated spots.
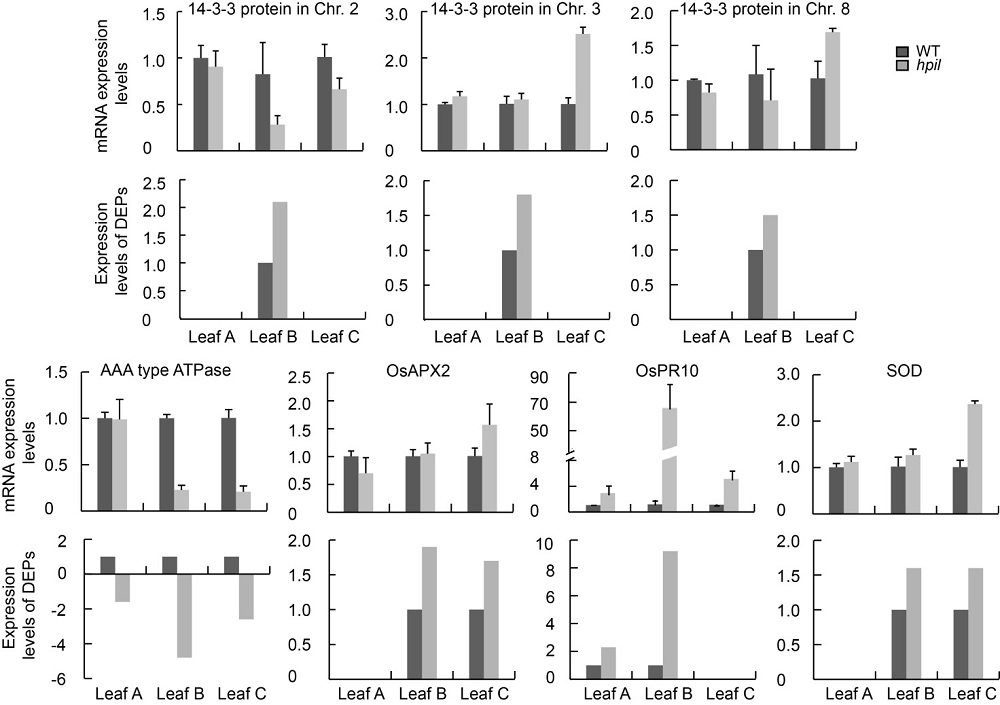
Fig. 3. Quantitative real-time PCR analysis of relative mRNA levels of differentially expressed proteins (DEPs) in leaves of WT and hpil.AAA type ATPase, ATPase associated with various cellular activities; SOD, Superoxide dismutase.WT, Zhejing 22; Leaf A, The first leaf from the top with no lesion mimic; Leaf B, The second leaf from the top with mild lesion mimic; Leaf C, The third leaf from the top with severe lesion mimic.Data are Mean ± SD (n = 3).
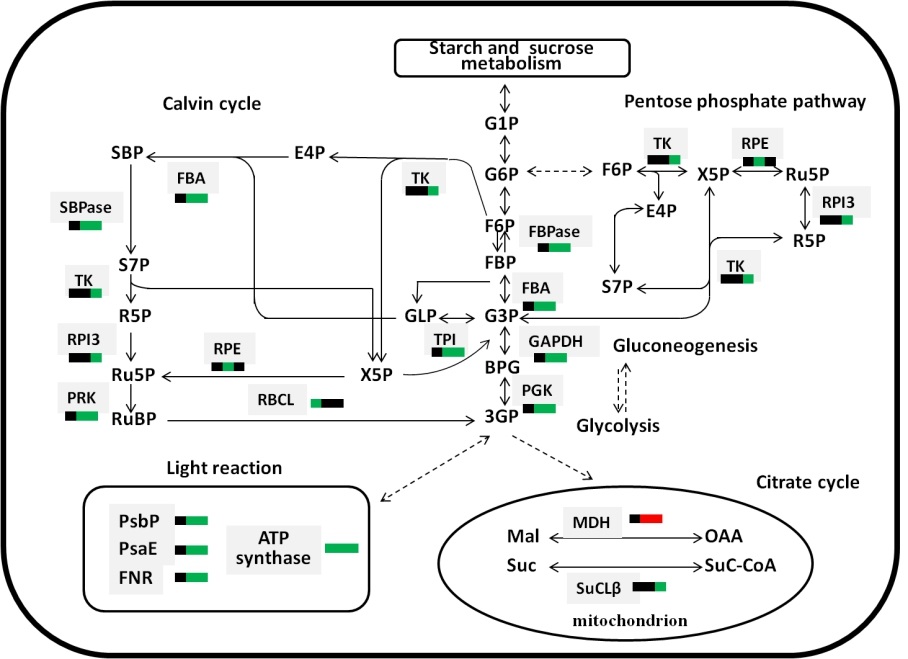
Fig. S5. Differently expressed proteins in the major metabolic pathways of rice.Three squares from left to right represent differential changes in leaf A, leaf B, leaf C in wild plant and hpil. Up-regulated (red), down-regulated (green), no change (black). The ratios of DEPs are listed in Table S2. SBPase (Sedoheptulose-1,7- bisphosphatase), TK (Transketolase), RPI 3 (Ribose 5-phosphate isomerase 3), PRK (Phosphoribulose kinase), RPE (Ribulosephosphate 3-epimerase), RBCL (Ribulose bisphosphate carboxylase large chain), FBA (Fructose bisphosphate aldolase), TPI (Triosephosphate isomerase), GAPDH (3-phosphate dehydrogenase), PGK (Phosphoglycerate kinase), FBPase (Fructose 1,6-bisphosphatase), MDH (Malate dehydrogenase), SuCLβ(Succinyl-CoA ligase beta-chain), S7P (Sedoheptulose 7-phosphate), R5P (Ribose 5 phosphate), Ru5P (Ribulose 5 phosphate), RUBP (Ribulose-1,5- bisphosphate), 3GP (glycerol-3-phoshpate), BPG (1,3 diphosphoglyceric acid), G3P (glyceraldehyde 3-phosphate), FBP (β-D Fructose 1,6-biphosphate), F6P (β-D Fructose 6-phosphate), G6P (α-D-Glucose 6-phosphate), G1P (α-D-Glucose 1 phosphate), E4P (Erythrose 4-phosphate), X5P (Xylulose 5-phosphate), OAA (Oxalaetic acid), Suc (Succinate), Mal (Malic acid).
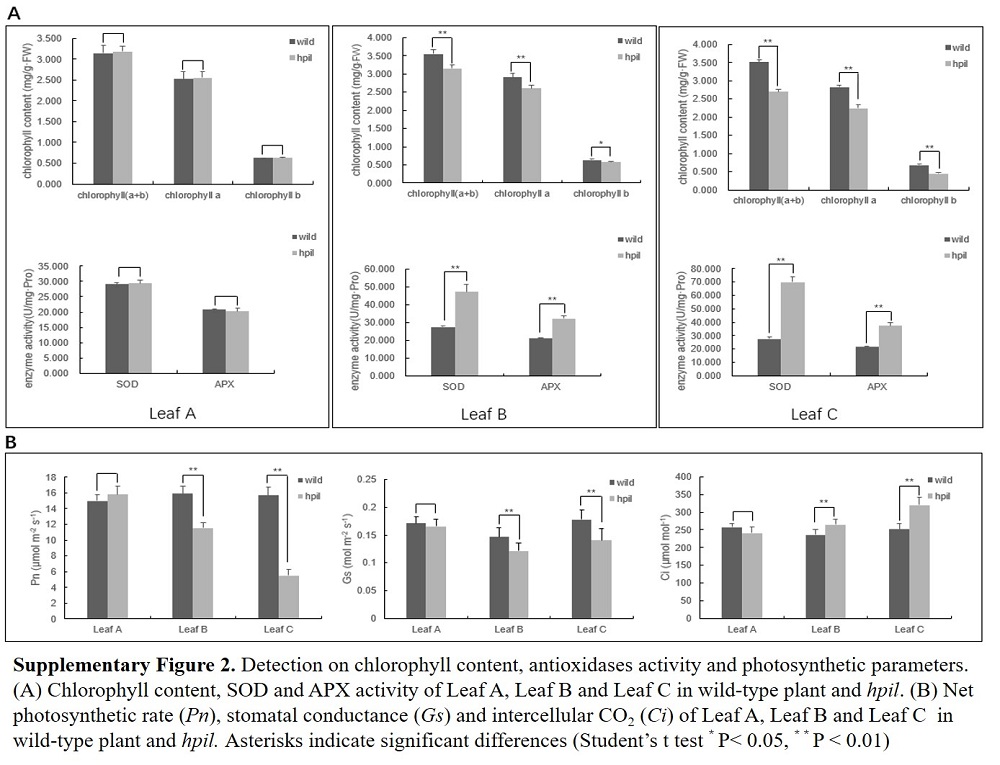
Fig. S6. Detection on chorophyll content, antioxidases activity and photosynthetic parameters.A, Chorophyll content, SOD and APX activity of leaf A, B and C in wild-type and hpil.B, Net photosynthetic rate (Pn), stomatal conductance (Gs) and intercellular CO2 (Ci) of leaf A, B and C in wild-type and hpil.Asterisks indicate significant differences (*, **, P < 0.05 and P < 0.01 by the Student’s t-test).
| [1] | Akhkha A, Clarke D D. 2003. Relative tolerances of wild and cultivated barleys to infection by Blumeria graminis f. sp. hordei (Syn. Erysiphe graminis f. sp. hordei): I. The effects of infection on growth and development. Physiol Mol Plant Pathol, 62(4): 237-250. |
| [2] | Anonymous.2002. Find out how the qualities of rice are evaluated and scored in this authoritative sourcebook. In: Standard Evaluation System for Rice. Laguna, the Philippines: International Rice Research Institute: 1518. |
| [3] | Arnon D I. 1949. Copper enzymes in isolated chloroplasts: Polyphenoloxidase in Beta vulagaris. Plant Physiol, 24(1): 1-15. |
| [4] | Beckman K B, Ames B N. 1997. Oxidative decay of DNA. J Biol Chem, 272(32): 19633‒19636. |
| [5] | Bowling S A, Clarke J D, Liu Y D, Klessig D F, Dong X. 1997. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell, 9(9): 1573-1584. |
| [6] | Bruggeman Q, Raynaud C, Benhamed M, Delarue M. 2015. To die or not to die? Lessons from lesion mimic mutants. Front Plant Sci, 6: 24. |
| [7] | Chen F, Li Q, Sun L X, He Z H. 2006. The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res, 13(2): 53-63. |
| [8] | Chen T, Chen Z, Sathe A P, Zhang Z H, Li L J, Shang H H, Tang S Q, Zhang X B, Wu J L. 2019. Characterization of a novel gain-of-function spotted-leaf mutant with enhanced disease resistance in rice. Rice Sci, 26(6): 372-383. |
| [9] | Cheng Y T, Li Y Z, Huang S, Huang Y, Dong X N, Zhang Y L, Li X. 2011. Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci USA, 108(35): 14694-14699. |
| [10] | Cheng Y T, Li X. 2012. Ubiquitination in NB-LRR-mediated immunity. Curr Opin Plant Biol, 15(4): 392-399. |
| [11] | Coll N S, Epple P, Dangl J L. 2011. Programmed cell death in the plant immune system. Cell Death Differ, 18(8): 1247-1256. |
| [12] | Corpas F J, González-Gordo S, Palma J M. 2020. Plant peroxisomes: A factory of reactive species. Front Plant Sci, 11: 853. |
| [13] | Crafts-Brandner S J, van de Loo F J, Salvucci M E. 1997. The two forms of ribulose-1,5-bisphosphate carboxylase/oxygenase activase differ in sensitivity to elevated temperature. Plant Physiol, 114(2): 439-444. |
| [14] | da S Andrade L B, Oliveira A S, Ribeiro J K C, Kiyota S, Vasconcelos I M, de Oliveira J T A, de Sales M P. 2010. Effects of a novel pathogenesis-related class 10 (PR-10) protein from Crotalaria pallida roots with papain inhibitory activity against root-knot nematode Meloidogyne incognita. J Agric Food Chem, 58(7): 4145-4152. |
| [15] | Dangl J L, Dietrich R A, Richberg M H. 1996. Death don’t have no mercy: Cell death programs in plant-icrobe interactions. Plant Cell, 8(10): 1793-1807. |
| [16] | Deng Z P, Zhang X, Tang W Q, Oses-Prieto J A, Suzuki N, Gendron J M, Chen H J, Guan S H, Chalkley R J, Peterman T K, Burlingame A L, Wang Z Y. 2007. A proteomics study of brassinosteroid response in Arabidopsis. Mol Cell Proteomics, 6(12): 2058-2071. |
| [17] | Dong Y, Fang X P, Yang Y, Xue G P, Chen X, Zhang W L, Wang X M, Yu C L, Zhou J, Mei Q, Fang W, Yan C Q, Chen J P. 2017. Comparative proteomic analysis of susceptible and resistant rice plants during early infestation by small brown planthopper. Front Plant Sci, 8: 1744. |
| [18] | Fang X P, Chen W Y, Xin Y, Zhang H M, Yan C Q, Yu H, Liu H, Xiao W F, Wang S Z, Zheng G Z, Liu H B, Jin L, Ma H S, Ruan S L. 2012. Proteomic analysis of strawberry leaves infected with Colletotrichum fragariae. J Proteomics, 75(13): 4074-4090. |
| [19] | Fekih R, Tamiru M, Kanzaki H, Abe A, Yoshida K, Kanzaki E, Saitoh H, Takaqi H, Natsume S, Undan J R, Undan J, Terauchi R. 2015. The rice (Oryza sativa L.) LESION MIMIC RESEMBLING, which encodes an AAA-type ATPase, is implicated in defense response. Mol Genet Genomics, 290(2): 611-622. |
| [20] | Flores T, Alape-Girón A, Flores-Díaz M, Flores H E. 2002. Ocatin: A novel tuber storage protein from the andean tuber crop oca with antibacterial and antifungal activities. Plant Physiol, 128(4): 1291-1302. |
| [21] | Giannopolitis C N, Ries S K. 1977. Superoxide dismutases: I. occurrence in higher plants. Plant Physiol, 59(2): 309-314. |
| [22] | Gordon T R, Duniway J M. 1982. Effects of powdery mildew infection on the efficiency of CO2 fixation and light utilization by sugar beet leaves. Plant Physiol, 69(1): 139-142. |
| [23] | Hashimoto M, Kisseleva L, Sawa S, Furukawa T, Komatsu S, Koshiba T. 2004. A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol, 45(5): 550-559. |
| [24] | Henkes S, Sonnewald U, Badur R, Flachmann R, Stitt M. 2001. A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell, 13(3): 535-552. |
| [25] | Hiraga S, Yamamoto K, Ito H, Sasaki K, Matsui H, Honma M, Nagamura Y, Sasaki T, Ohashi Y. 2000. Diverse expression profiles of 21 rice peroxidase genes. FEBS Lett, 471: 245‒250. |
| [26] | Huang L F, Lin K H, He S L, Chen J L, Jiang J Z, Chen B H, Hou Y S, Chen R S, Hong C Y, Ho S L. 2016. Multiple patterns of regulation and overexpression of a ribonuclease-like pathogenesis- related protein gene,OsPR10a, conferring disease resistance in rice and Arabidopsis. PLoS One, 11(6): e0156414. |
| [27] | Huang Q N, Yang Y, Shi Y F, Chen J, Wu J L. 2010. Spotted-leaf mutants of rice (Oryza sativa). Rice Sci, 17(4): 247-256. |
| [28] | Jones J D G, Dangl J L. 2006. The plant immune system. Nature, 444: 323‒329. |
| [29] | Jwa N S, Agrawal G K, Rakwal R, Park C H, Agrawal V P. 2001. Molecular cloning and characterization of a novel jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem Biophy Res Commun, 286(5): 973-983. |
| [30] | Kang S G, Matin M N, Bae H, Natarajan S. 2007. Proteome analysis and characterization of phenotypes of lesion mimic mutant spotted leaf 6 in rice. Proteomics, 7(14): 2447-2458. |
| [31] | Kauffman H E, Reddy A P K, Hsieh S P Y, Merca S D. 1973. An improved technique for 436 evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep, 57: 737‒741. |
| [32] | Liu Q E, Ning Y S, Zhang Y X, Yu N, Zhao C D, Zhan X D, Wu W X, Chen D B, Wei X J, Wang G L, Cheng S H, Cao L Y. 2017. OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell, 29(2): 345-359. |
| [33] | Lorrain S, Vailleau F, Balagué C, Roby D. 2003. Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci, 8(6): 263-271. |
| [34] | Lozano-Durán R, Robatzek S. 2015. 14-3-3 proteins in plant- pathogen interactions. Mol Plant Microbe Interact, 28(5): 511-518. |
| [35] | Manosalva P M, Bruce M, Leach J E. 2011. Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance. Plant J, 68(5): 777-787. |
| [36] | Mcgee J D, Hamer J E, Hodges T K. 2001. Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Mol Plant Microbe Interact, 14(7): 877-886. |
| [37] | Mei Q, Yang Y, Ye S H, Liang W F, Wang X M, Zhou J, Yu C L, Yan C Q, Chen J P. 2019. H2O2 induces association of RCA with the thylakoid membrane to enhance resistance of Oryza meyeriana to Xanthomonas oryzae pv. oryzae. Plants, 8(9): 351. |
| [38] | Nakano Y, Asada K. 1981. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol, 22(5): 867-880. |
| [39] | Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones J D G. 2004. The transcriptional innate immune response to flg22: Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol, 135(2): 1113-1128. |
| [40] | Park C J, Kim K J, Shin R, Park J M, Shin Y C, Peak K H. 2004. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J, 37(2): 186-198. |
| [41] | Qin P, Fan S J, Deng L C, Zhong G R, Zhang S W, Li M, Chen W L, Wang G L, Tu B, Wang Y P, Chen X W, Ma B T, Li S G. 2018. LML1, encoding a conserved eukaryotic release factor 1 protein, regulates cell death and pathogen resistance by forming a conserved complex with SPL33 in rice. Plant Cell Physiol, 59(5): 887-902. |
| [42] | Rodriguez E, Ghoul H E, Mundy J, Petersen M. 2015. Making sense of plant autoimmunity and ‘negative regulators’. FEBS J, 283(8): 1385-1391. |
| [43] | Sharkey T D, Badger M R, von Caemmerer S, Andrews T J. 2001. Increased heat sensitivity of photosynthesis in tobacco plants with reduced Rubisco activase. Photosynth Res, 67(1): 147-156. |
| [44] | Shen X L, Liu H B, Yuan B, Li X H, Xu C G, Wang S P. 2011. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ, 34(2): 179-191. |
| [45] | Suzuki N, Miller G, Morales J, Shulaev V, Torres M A, Mittler R. 2011. Respiratory burst oxidases: The engines of ROS signaling. Curr Opin Plant Biol, 14(6): 691-699. |
| [46] | Tang J Y, Zhu X D, Wang Y Q, Liu L C, Xu B, Li F, Fang J, Chu C C. 2011. Semi-dominant mutations in the CC-NB-LRR-type R gene, NLS1, lead to constitutive activation of defense responses in rice. Plant J, 66(6): 996-1007. |
| [47] | Thordal-Christensen H, Zhang Z G, Wei Y D, Collinge D B. 1997. Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J, 11(6): 1187-1194. |
| [48] | Tsuda K, Katagiri F. 2010. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol, 13(4): 459-465. |
| [49] | van Loon L C, van Strien E A. 1999. The families of pathogenesis- related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol, 55(2): 85-97. |
| [50] | Wang L J, Pei Z Y, Tian Y C, He C Z. 2005. OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol Plant Microbe Interact, 18(5): 375-384. |
| [51] | Wang S, Lei C L, Wang J L, Ma J, Tang S, Wang C L, Zhao K J, Tian P, Zhang H, Qi C Y, Cheng Z J, Zhang X, Guo X P, Liu L L, Wu C Y, Wan J M. 2017. SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J Exp Bot, 68(5): 899-913. |
| [52] | Wang X C, Wang D Y, Wang D, Wang H Y, Chang L L, Yi X P, Peng M, Guo A P. 2012. Systematic comparison of technical details in CBB methods and development of a sensitive GAP stain for comparative proteomic analysis. Electrophoresis, 33(2): 296-306. |
| [53] | Wang Z H, Wang Y, Xiao H, Hu D H, Liu C X, Yang J, Li Yang, Huang Y Q, Feng Y Q, Gong H Y, Li Y, Fang G, Tang H R, Li Y S. 2015. Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J Exp Bot, 66(3): 973-987. |
| [54] | Waszczak C, Carmody M, Kangasjärvi J. 2018. Reactive oxygen species in plant signaling. Annu Rev Plant Biol, 69: 209-236. |
| [55] | Wu J N, Kim S G, Kang KY, Kim J G, Park S R, Gupta R, Kim Y H, Wang Y M, Kim S T. 2016. Overexpression of a pathogenesis- related protein 10 enhances biotic and abiotic stress tolerance in rice. Plant Pathol J, 32(6): 552-562. |
| [56] | Xia Z L, Wei Y Y, Sun K L, Wu J Y, Wang Y X, Wu K. 2013. The maize AAA-type protein SKD1 confers enhanced salt and drought stress tolerance in transgenic tobacco by interacting with lyst-interacting protein 5. PLoS One, 8(7): e69787. |
| [57] | Xie Y R, Chen Z Y, Brown R L, Bhatnagar D. 2010. Expression and functional characterization of two pathogenesis-related protein 10 genes from Zea mays. J Plant Physiol, 167(2): 121-130. |
| [58] | Yan S P, Zhang Q Y, Tang Z C, Su W A, Sun W N. 2005. Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteomics, 5(3): 484-496. |
| [59] | Yin Z, Chen J, Zeng L, Goh M, Leung H, Khush G S, Wang G L. 2000. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol Plant Microbe Interact, 13(8): 869-876. |
| [60] | Zeng L R, Qu S H, Bordeos A, Yang C W, Baraoidan M, Yan H Y, Xie Q, Nahm B H, Leung H, Wang G L. 2004. Spotted leaf 11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell, 16(10): 2795-2808. |
| [61] | Zhang H F, Tang W, Liu K Y, Huang Q, Zhang X, Yan X, Chen Y, Wang J S, Qi Z Q, Wang Z Y, Zheng X B, Wang P, Zhang Z G. 2011. Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog, 7(12): e1002450. |
| [62] | Zhang X B, Feng B H, Wang H M, Xu X, Shi Y F, He Y, Chen Z, Sathe A P, Shi L, Wu J L. 2018. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J Integr Plant Biol, 60(2): 160-172. |
| [63] | Zhao J Y, Liu P C, Li C R, Wang Y Y, Guo L Q, Jiang G H, Zhai W X. 2017. LMM5.1 and LMM5.4, two eukaryotic translation elongation factor 1A-like gene family members, negatively affect cell death and disease resistance in rice. J Genet Genomics, 44(2): 107-118. |
| [64] | Zhou L, Cheung M Y, Li M W, Fu Y P, Sun Z X, Sun S M, Lam M H. 2010. Rice hypersensitive induced reaction protein 1 (OsHIR1) associates with plasma membrane and triggers hypersensitive cell death. BMC Plant Biol, 10(1): 290. |
| [65] | Zhu X B, Mu Z, Mawsheng C, Chen X W, Wang J. 2020. Deciphering rice lesion mimic mutants to understand molecular network governing plant immunity and growth. Rice Sci, 27(4): 278-288. |
| [1] | Mishra Rukmini, Zheng Wei, Kumar Joshi Raj, Kaijun Zhao. Genome Editing Strategies Towards Enhancement of Rice Disease Resistance [J]. Rice Science, 2021, 28(2): 133-145. |
| [2] | Yanchang Luo, Tingchen Ma, Teo Joanne, Zhixiang Luo, Zefu Li, Jianbo Yang, Zhongchao Yin. Marker-Assisted Breeding of Thermo-Sensitive Genic Male Sterile Line 1892S for Disease Resistance and Submergence Tolerance [J]. Rice Science, 2021, 28(1): 89-98. |
| [3] | Xiaobo Zhu, Mu Ze, Mawsheng Chern, Xuewei Chen, Jing Wang. Deciphering Rice Lesion Mimic Mutants to Understand Molecular Network Governing Plant Immunity and Growth [J]. Rice Science, 2020, 27(4): 278-288. |
| [4] | B. ANGELES-SHIM Rosalyn, P. REYES Vincent, M. del VALLE Marilyn, S. LAPIS Ruby, SHIM Junghyun, SUNOHARA Hidehiko, K. JENA Kshirod, ASHIKARI Motoyuki, DOI Kazuyuki. Marker-Assisted Introgression of Quantitative Resistance Gene pi21 Confers Broad Spectrum Resistance to Rice Blast [J]. Rice Science, 2020, 27(2): 113-123. |
| [5] | Ting Chen, Zheng Chen, Prakash Sathe Atul, Zhihong Zhang, Liangjian Li, Huihui Shang, Shaoqing Tang, Xiaobo Zhang, Jianli Wu. Characterization of a Novel Gain-of-Function Spotted-Leaf Mutant with Enhanced Disease Resistance in Rice [J]. Rice Science, 2019, 26(6): 372-383. |
| [6] | Jiehua Qiu, Shuai Meng, Yizhen Deng, Shiwen Huang, Yanjun Kou. Ustilaginoidea virens: A Fungus Infects Rice Flower and Threats World Rice Production [J]. Rice Science, 2019, 26(4): 199-206. |
| [7] | Tao Chen, Hao Wu, Ya-dong Zhang, Zhen Zhu, Qi-yong Zhao, Li-hui Zhou, Shu Yao, Ling Zhao, Xin Yu, Chun-fang Zhao, Cai-lin Wang. Genetic Improvement of Japonica Rice Variety Wuyujing 3 for Stripe Disease Resistance and Eating Quality by Pyramiding Stv-bi and Wx-mq [J]. Rice Science, 2016, 23(2): 69-77. |
| [8] | MA Jian-yang1, 2, CHEN Sun-lu2, 3, ZHANG Jian-hui1, 2, DONG Yan-jun1, TENG Sheng2. Identification and Genetic Mapping of a Lesion Mimic Mutant in Rice [J]. RICE SCIENCE, 2012, 19(1): 1-7. |
| [9] | YAO Shu1, 2, CHEN Tao1, ZHANG Ya-dong1, ZHU Zhen1, ZHAO Ling1, ZHAO Qing-yong1, . Transferring Translucent Endosperm Mutant Gene Wx-mq and Rice Stripe Disease Resistance Gene Stv-bi by Marker-Assisted Selection in Rice (Oryza sativa) [J]. RICE SCIENCE, 2011, 18(2): 102-109. |
| [10] | HUANG Qi-na, YANG Yang, SHI Yong-feng, CHEN Jie, WU Jian-li. Spotted-Leaf Mutants of Rice (Oryza sativa) [J]. RICE SCIENCE, 2010, 17(4): 247-256 . |
| [11] | CHEN De-xi, CHEN Xue-wei, LEI Cai-lin, MA Bing-tian, WANG Yu-ping, LI Shi-gui. Rice Blast Resistance of Transgenic Rice Plants with Pi-d2 Gene [J]. RICE SCIENCE, 2010, 17(3): 179-184 . |
| [12] | WANG Zhong-hua, JIA Yu-lin, LIN Hui, Adair INTERN, Barbara VALENT, J. Neil RUTGER . Host Active Defense Responses Occur within 24 Hours after Pathogen Inoculation in the Rice Blast System [J]. RICE SCIENCE, 2007, 14(4): 302-310 . |
| [13] | CAO Li-yong, ZHUANG Jie-yun, YUAN Shou-jiang, ZHAN Xiao-deng, ZHENG Kang-le, CHENG Shi-hua. Hybrid Rice Resistant to Bacterial Leaf Blight Developed By Marker Assisted Selection [J]. RICE SCIENCE, 2003, 11(1-2): 68-70 . |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||