
Rice Science ›› 2016, Vol. 23 ›› Issue (3): 152-159.DOI: 10.1016/j.rsci.2016.04.002
收稿日期:2015-08-27
接受日期:2015-12-09
出版日期:2016-06-08
发布日期:2016-02-04
. [J]. Rice Science, 2016, 23(3): 152-159.
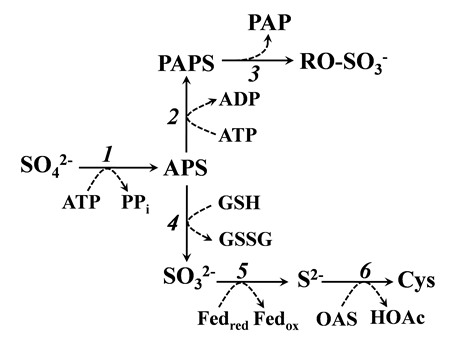
Fig. 1. Scheme of sulfate assimilation pathway in plant chloroplast.Dotted arrows indicated the cosubstrates and by-products. APS, Adenosine 5'-phosphosulfate; PAPS, 3'-phospho-adenosine- 5'-phosphosulfate; PAP, 3',5'-bisphosphate adenosine; RO-SO3-, Sulfated compounds; GSH, Reduced glutathione; GSSG, Oxidized glutathione; Fedred, Reduced thioredoxin; Fedox, Oxidized thioredoxin; OAS, O-acetylserine; HOAc, Acetate; Cys, Cysteine; 1, ATP sulfurylase; 2, APS kinase; 3, Sulfotransferase; 4, APS reductase; 5, Sulfite reductase; 6, O-acetylserine sulfhydrylase.
| Primer | Sequence (5'-3') |
|---|---|
| OsAPSK1-F | GAATTCATGGAGCAGCAGCAGC |
| OsAPSK1-R | CTCGAGTTAAGCTTGCAAATAT |
| APSK1-C36A-F | CAATATACTGTGGCACAATGCGCCAATTGGACAATCTG |
| APSK1-C36A-R | CAGATTGTCCAATTGGCGCATTGTGCCACAGTATATTG |
| APSK1-C69A-F | GGGAAAAGCACTCTTGCAGCGGCACTGAATCGGGAG |
| APSK1-C69A-R | CTCCCGATTCAGTGCCGCTGCAAGAGTGCTTTTCCC |
Table 1 Primers for gene cloning and mutagenesis.
| Primer | Sequence (5'-3') |
|---|---|
| OsAPSK1-F | GAATTCATGGAGCAGCAGCAGC |
| OsAPSK1-R | CTCGAGTTAAGCTTGCAAATAT |
| APSK1-C36A-F | CAATATACTGTGGCACAATGCGCCAATTGGACAATCTG |
| APSK1-C36A-R | CAGATTGTCCAATTGGCGCATTGTGCCACAGTATATTG |
| APSK1-C69A-F | GGGAAAAGCACTCTTGCAGCGGCACTGAATCGGGAG |
| APSK1-C69A-R | CTCCCGATTCAGTGCCGCTGCAAGAGTGCTTTTCCC |
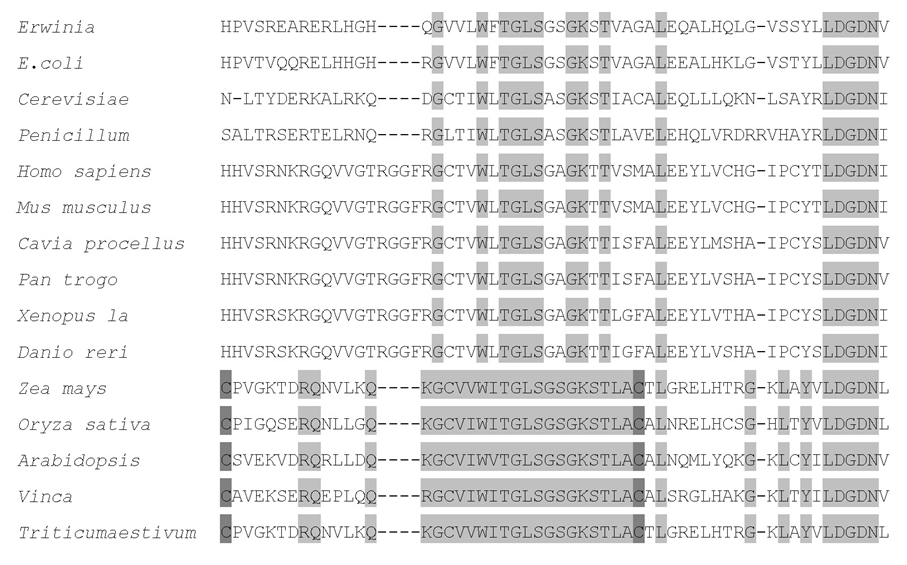
Fig. 2. Sequence alignment of adenosine 5′-phosphosulfate kinase (APSKs) from different species.Conserved amino acids are highlighted as gray background, and conserved cysteines in plant APSKs are highlighted as dark gray background.
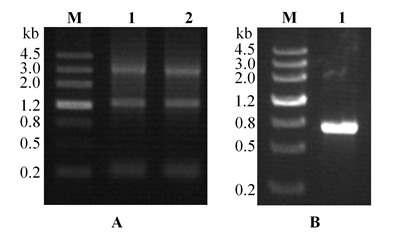
Fig. 3. Gene cloning of OsAPSK1. A, Total mRNA from rice leaves. M, Nucleotide marker (kb); Lanes 1 and 2, mRNA (total RNA) extracted from rice leaves. B, reverse transcription PCR amplified OsAPSK1 fragment. M, Nucleotide marker (kb); Lane 1, OsAPSK1 nucleotide fragment (720 bp).
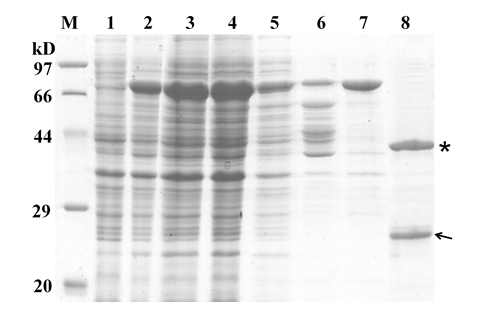
Fig. 4. Expression and purification of OsAPSK1. M, Protein molecular marker (kD); Lane 1, Before isopropyl β-D-1-thiogalactopyranoside (IPTG) induction; Lane 2, After IPTG induction; Lane 3, Supernatant of cell lysate after centrifugation; Lane 4, Lysis buffer washing; Lane 5, High salt washing; Lane 6, Low salt washing; Lane 7, Elution of fusion protein HAL2-APSK; Lane 8, Cleavage of HAL2-APSK by prescission protease (HAL2 tag and OsAPSK1 were indicated by star and arrow, respectively).
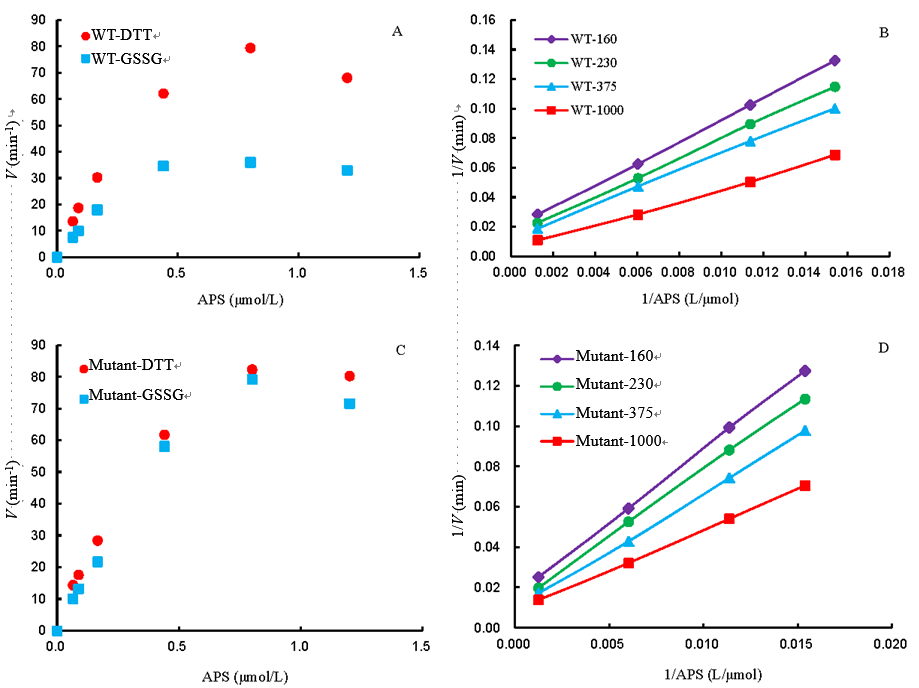
Fig. 5. Initial rate assays for phosphorylation of adenosine 5-phosphosulfate (APS) by wild type OsAPSK1 and C36A/C69A OsAPSK1. A, Activity comparison at different concentrations of APS in presence of dithiothreitol (DTT) (1.0 mmol/L) or oxidized glutathione (GSSG) (0.5 mmol/L) by wild type OsAPSK1; B, Double reciprocal plot for APS phosphorylation activity by wild type OsAPSK1 with ATP concentrations of 160, 230, 375 and 1 000 μmol/L, respectively; C, Activity comparison at different concentrations of APS in presence of DTT (1.0 mmol/L) or GSSG (0.5 mmol/L) by C36A/C69A OsAPSK1; D, Double reciprocal plot for APS phosphorylation activity by C36A/C69A OsAPSK1 with ATP concentrations of 160, 230, 375 and 1 000 μmol/L, respectively.
| Parameter | KmAPS | KmATP | KiATP | KiAPS | kcat | kcat/KmAPS |
|---|---|---|---|---|---|---|
| (μmol/L) | (mmol/L) | (mmol/L) | (μmol/L) | (min-1) | (L/μmol∙min) | |
| Wild type | 0.72 ± 0.17 | 0.75 ± 0.18 | 0.61 ± 0.13 | 0.58 ± 0.13 | 300 ± 42 | 417 |
| C36A/C69A | 0.44 ± 0.04 | 0.14 ± 0.02 | 0.22 ± 0.04 | 0.68 ± 0.15 | 130 ± 6 | 295 |
Table 2 Comparison of kinetic parameters for wild type OsAPSK1 and mutant C36A/C69A OsAPSK1.
| Parameter | KmAPS | KmATP | KiATP | KiAPS | kcat | kcat/KmAPS |
|---|---|---|---|---|---|---|
| (μmol/L) | (mmol/L) | (mmol/L) | (μmol/L) | (min-1) | (L/μmol∙min) | |
| Wild type | 0.72 ± 0.17 | 0.75 ± 0.18 | 0.61 ± 0.13 | 0.58 ± 0.13 | 300 ± 42 | 417 |
| C36A/C69A | 0.44 ± 0.04 | 0.14 ± 0.02 | 0.22 ± 0.04 | 0.68 ± 0.15 | 130 ± 6 | 295 |

Fig. 6. Structural illustration of OsAPSK1. A, Dimeric OsAPSK1. Blue and green cartoon models are monomers of OsAPSK1. Arrows point to sphere model of Cysteine 36 (C36) and Cysteine 69 (C69) from different monomers; B, Interactions between C36 and C69 in detail. Arrows point to sticks model of C36 and C69. The distance between sulfur atoms from C36 and C69 is 2.1Å.
| [1] | Arnold K, Bordoli L, Kopp J, Schwede T.2006. The SWISS- MODEL workspace: A web-based environment for protein structure homology modelling.Bioinformatics, 22(2): 195-201. |
| [2] | Cleland W W.1979. Statistical analysis of enzyme kinetic data.Methods Enzymol, 63(2): 103-138. |
| [3] | Dong T.2010. Functional analysis of rice sulfate transporter gene OsSultr1;1. Ji’nan: Shandong University. (in Chinese with English abstract) |
| [4] | Hermann M, Maier F, Masroor A, Hirth S, Pfitzner A J P, Pfitzner U M.2013. The Arabidopsis NIMIN proteins affect NPR1 differentially. Front Plant Sci, 4: 1-15. |
| [5] | Kumar V, Joshi S G, Bell A A, Rathore K S.2013. Enhanced resistance against Thielaviopsis basicola in transgenic cotton plants expressing Arabidopsis NPR1 gene.Transg Res, 22(2): 359-368. |
| [6] | Lee B R, Huseby S, Koprivova A, Chetelat A, Wirtz M, Mugford S T, Navid E, Brearley C, Saha S, Mithen R, Hell R, Farmer E E, Kopriva S.2012. Effects of fou8/fry1 mutation on sulfur metabolism: Is decreased internal sulfate the trigger of sulfate starvation response?PLoS One, 7(6): e39425. |
| [7] | Leyh T S.1993. The physical biochemistry and molecular genetics of sulfate activation.Crit Rev Biochem Mol Biol, 28(6): 515-542. |
| [8] | Li H Q, Li Z, Wang Y Y, Liu X, Wang Q G, Yao F Y, Liu W.2013. Transformation of γ-thionin gene RsAFP1 into the rice and preliminary identification of its blast resistance.Chin J Rice Sci, 27(4): 335-343. (in Chinese with English abstract) |
| [9] | Li H Y, Zhou S T, Ma J H, Sun M H.2013. Purification and characterization of pyruvate kinase II from Escherichia coli and its application as a coupling enzyme.China Biotechnol, 33(5): 68-74. (in Chinese with English abstract) |
| [10] | Li X J, Xu D D, Xu Y M, Zhai K E, Yang Y L, Pan J W, Rao Y C.2015. Expression of OsBC88, a rice cellulose synthase catalytic subunit gene.Chin J Rice Sci, 29(2): 126-134. (in Chinese with English abstract) |
| [11] | Liu J.2007. Characteristic of adenosine sulfurylase and 3'-phospho- adenosine-5'-phospho-sulfate reductase genes in rice. Ji’nan: Shandong University. (in Chinese with English abstract) |
| [12] | Mueller J W, Shafqat N.2013. Adenosine 5'-phosphosulfate: A multifaceted modulator of bifunctional 3'-phosphoadenosine-5'- phosphosulfate synthases and related enzymes.FEBS J, 280(13): 3050-3057. |
| [13] | Mugford S G, Lee B R, Koprivova A, Matthewman C, Kopriva S.2011. Control of sulfur partitioning between primary and secondary metabolism. Plant J, 65(1): 96-105. |
| [14] | Mugford S G, Yoshimoto N, Reichelt M, Wirtz M, Hill L, Mugford S T, Nakazato Y, Noji M, Takahashi H, Kramell R, Gigolashvili T, Flugge U I, Wasternack C, Gershenzon J, Hell R, Saito K, Kopriva S.2009. Disruption of adenosine 5'-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites.Plant Cell, 21(3): 910-927. |
| [15] | Ravilious G E, Jez J M.2012. Nucleotide binding site communication in Arabidopsis thaliana adenosine 5'-phosphosulfate kinase. J Biol Chem, 287(36): 30385-30394. |
| [16] | Ravilious G E, Nguyen A, Francois J A, Jez J M.2012. Structural basis and evolution of redox regulation in plant adenosine 5'-phosphosulfate kinase.Proc Natl Acad Sci USA, 109(1): 309-314. |
| [17] | Ravilious G E, Westfall C S, Jez J M.2013. Redox-linked gating of nucleotide binding by the N-terminal domain of adenosine 5'-phosphosulfate kinase.J Biol Chem, 288(9): 6107-6115. |
| [18] | Ryu S E.2012. Structural mechanism of disulphide bond-mediated redox switches.J Biochem, 151(6): 579-588. |
| [19] | Saito K.2004. Sulfur assimilatory metabolism: The long and smelling road.Plant Physiol, 136(1): 2443-2450. |
| [20] | Song Y E, Wang X, Shen Z W, Xu Y, Li J Y.2013. Expressing the maize anthocyanin regulatory gene Lc increased flavonoid content in the seed of white pericarp rice and purple pericarp rice.Genetika, 49(11): 1292-1299. |
| [21] | Sun M H, Andreassi J L, Liu S Q, Pinto R, Triccas J A, Leyh T S.2005. The trifunctional sulfate-activating complex (SAC) of Mycobacterium tuberculosis.J Biol Chem, 280(9): 7861-7866. |
| [22] | Sun X M, Yang Z M.2006. Plant sulfate assimilation and regulation of the activity of related enzymes under cadmium stress. J Plant Physiol Mol Biol, 32(1): 9-16. (in Chinese with English abstract) |
| [23] | Villanueva-Alonzo H J, Us-Camas R Y, Lopez-Ochoa L A, Robertson D, Guerra-Peraza O, Minero-Garcia Y, Moreno- Valenzuela O A.2013. A new virus-induced gene silencing vector based on Euphorbia mosaic virus-Yucatan peninsula for NPR1 silencing in Nicotiana benthamiana and Capsicum annuum var. Anaheim.Biotechnol Lett, 35(5): 811-823. |
| [24] | Wang Y X, Yang L, Sun M H.2011. Classification and functions of sulfate activating complexes.Chem Life, 31(2): 252-257. (in Chinese with English abstract) |
| [25] | Wei J, Tang Q X, Varlamova O, Roche C, Lee R, Leyh T S.2002. Cysteine biosynthetic enzymes are the pieces of a metabolic energy pump.Biochemistry, 41(26): 8493-8498. |
| [26] | Xing F S, Song P, Wang F T, Gao Y Z.1992. Response of glutathione-ascorbate cycle in rice leaves to photoinhibition.Chin J Rice Sci, 6(4): 177-183. (in Chinese with English abstract) |
| [27] | Yang Y, Ma J H, Yang Y L, Zhang X, Wang Y X, Yang L, Sun M H.2014. Yeast 3',5'-bisphosphate nucleotidase: An affinity tag for protein purification.Protein Exp Purif, 97: 81-87. |
| [28] | Zhu F Y, Chen Y Z, Yan X F.2007. Plant glucosinolate metabolism and sulfur nutrition.Plant Physiol Commun, 34(6): 1189-1195. (in Chinese with English abstract) |
| [1] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages[J]. Rice Science, 2023, 30(6): 7-. |
| [2] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage[J]. Rice Science, 2023, 30(6): 12-. |
| [3] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis[J]. Rice Science, 2023, 30(6): 8-. |
| [4] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines[J]. Rice Science, 2023, 30(6): 6-. |
| [5] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice[J]. Rice Science, 2023, 30(6): 4-. |
| [6] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications[J]. Rice Science, 2023, 30(6): 9-. |
| [7] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage[J]. Rice Science, 2023, 30(6): 14-. |
| [8] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance[J]. Rice Science, 2023, 30(6): 10-. |
| [9] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review[J]. Rice Science, 2023, 30(6): 5-. |
| [10] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats[J]. Rice Science, 2023, 30(6): 11-. |
| [11] | . [J]. Rice Science, 2021, 28(3): 217-232. |
| [12] | . [J]. Rice Science, 2019, 26(2): 118-124. |
| [13] | . [J]. Rice Science, 2019, 26(2): 77-87. |
| [14] | . [J]. Rice Science, 2019, 26(2): 88-97. |
| [15] | . [J]. Rice Science, 2019, 26(2): 98-108. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||