
Rice Science ›› 2015, Vol. 22 ›› Issue (5): 207-216.DOI: 10.1016/S1672-6308(14)60303-6
• Orginal Article • Next Articles
Lian-ping Sun1,2, Ying-xin Zhang1, Pei-pei Zhang1, Zheng-fu Yang1, Xiao-deng Zhan1, Xi-hong Shen1, Zhen-hua Zhang1, Xia Hu1, Dan-dan Xuan1, Wei-xun Wu1, Zi-he Li1, Li-yong Cao1( ), Shi-hua Cheng1,2(
), Shi-hua Cheng1,2( )
)
Received:2015-05-19
Accepted:2015-07-01
Online:2015-05-15
Published:2015-07-24
Lian-ping Sun, Ying-xin Zhang, Pei-pei Zhang, Zheng-fu Yang, Xiao-deng Zhan, Xi-hong Shen, Zhen-hua Zhang, Xia Hu, Dan-dan Xuan, Wei-xun Wu, Zi-he Li, Li-yong Cao, Shi-hua Cheng. K-Domain Splicing Factor OsMADS1 Regulates Open Hull Male Sterility in Rice[J]. Rice Science, 2015, 22(5): 207-216.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.org/EN/10.1016/S1672-6308(14)60303-6
| Primer | Forward primer (5′-3′) | Reverse primer (5′-3′) | Purpose |
|---|---|---|---|
| RM7576 | CTGCCCTGCCTTTTGTACAC | GCGAGCATTCTTTCTTCCAC | Linkage analysis |
| RM7 | TTCGCCATGAAGTCTCTCG | CCTCCCATCATTTCGTTGTT | Linkage analysis |
| InD44 | GGAATCCCTCCCTTCTTGTC | GGTCGGTAAAGACGGTGAAA | Fine mapping |
| InD45 | CCAGGGATCTTCTCATCCAA | CCTGGCTAGCATACCACACA | Fine mapping |
| InD46 | GCCATTGATCTTCTGCAGGT | TTTGTTGTCAATGCCCTGTT | Fine mapping |
| RD0304 | GGCGTCACTGCTCGTA | GCCTGAAGCGTCCACA | Fine mapping |
| KY2 | GTGGGAAGAAGAACATCAACTG | GCACACAAGATAAACCCAATCAGC | Fine mapping |
| KY12 | ACCACGAGGGTGACCGTAGA | GCGAGGGTTGATGAGATAGCA | Fine mapping |
| KY17 | CGAGAGGCGAAGGAAATAGAACG | CTCCTCCTCCTCCTGGTTCTCC | Fine mapping |
| KY25 | CCATGGTCGCCATTGACACG | CCTGCTATAACACTCGCACAGATGC | Fine mapping |
| KY26 | GGTGGTGAGCCAAGAACTGACC | CCTCAAGGAATCCTCGTAAGTCG | Fine mapping |
| KY29 | CCAAGTGTGTCCGAGCTTAGTGC | TGAGTCAAAGCGAAAGTCAACAGG | Fine mapping |
| CAPS1 | GCCATCGATCACCCTGAAAGTC | CTGATCAGCAAGAACAGTGC | ohms1 site detection |
| CKY | AGCCAAACCACACCACCATAAAG | AGGACACTGTTTGCATTGGCT | cDNA sequencing |
Table 1 Primer sequences used in this study.
| Primer | Forward primer (5′-3′) | Reverse primer (5′-3′) | Purpose |
|---|---|---|---|
| RM7576 | CTGCCCTGCCTTTTGTACAC | GCGAGCATTCTTTCTTCCAC | Linkage analysis |
| RM7 | TTCGCCATGAAGTCTCTCG | CCTCCCATCATTTCGTTGTT | Linkage analysis |
| InD44 | GGAATCCCTCCCTTCTTGTC | GGTCGGTAAAGACGGTGAAA | Fine mapping |
| InD45 | CCAGGGATCTTCTCATCCAA | CCTGGCTAGCATACCACACA | Fine mapping |
| InD46 | GCCATTGATCTTCTGCAGGT | TTTGTTGTCAATGCCCTGTT | Fine mapping |
| RD0304 | GGCGTCACTGCTCGTA | GCCTGAAGCGTCCACA | Fine mapping |
| KY2 | GTGGGAAGAAGAACATCAACTG | GCACACAAGATAAACCCAATCAGC | Fine mapping |
| KY12 | ACCACGAGGGTGACCGTAGA | GCGAGGGTTGATGAGATAGCA | Fine mapping |
| KY17 | CGAGAGGCGAAGGAAATAGAACG | CTCCTCCTCCTCCTGGTTCTCC | Fine mapping |
| KY25 | CCATGGTCGCCATTGACACG | CCTGCTATAACACTCGCACAGATGC | Fine mapping |
| KY26 | GGTGGTGAGCCAAGAACTGACC | CCTCAAGGAATCCTCGTAAGTCG | Fine mapping |
| KY29 | CCAAGTGTGTCCGAGCTTAGTGC | TGAGTCAAAGCGAAAGTCAACAGG | Fine mapping |
| CAPS1 | GCCATCGATCACCCTGAAAGTC | CTGATCAGCAAGAACAGTGC | ohms1 site detection |
| CKY | AGCCAAACCACACCACCATAAAG | AGGACACTGTTTGCATTGGCT | cDNA sequencing |
| Combination | Seed-setting rate of F1 | F2 | χ2(3:1) | χ20.05 | |
|---|---|---|---|---|---|
| No. of wild type plants | No. of mutant plants | ||||
| ohms1/ZH11 | 85.22 | 374 | 113 | 0.84 | 3.84 |
| ohms1/02428 | 88.76 | 4658 | 1496 | 1.57 | |
Table 2 Segregation analysis of ohms1 allele.
| Combination | Seed-setting rate of F1 | F2 | χ2(3:1) | χ20.05 | |
|---|---|---|---|---|---|
| No. of wild type plants | No. of mutant plants | ||||
| ohms1/ZH11 | 85.22 | 374 | 113 | 0.84 | 3.84 |
| ohms1/02428 | 88.76 | 4658 | 1496 | 1.57 | |
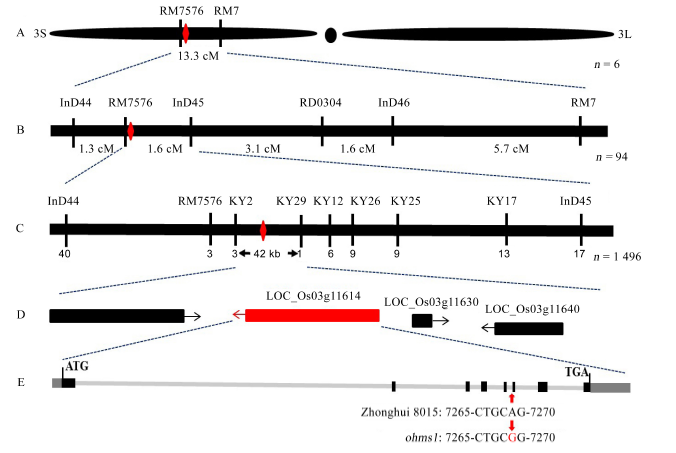
Fig. 2. Fine mapping and positional cloning of ohms1 mutant gene.(A, The mutate gene was linked between RM7576 and RM7 on the short arm of chromosome 3; B, The mutant gene was preliminarily mapped between RM7576 and InD45; C, The ohms1 mutant gene was fine-mapped to a 42-kb genomic region between markers KY2 and KY29; D, Four open reading frames existed in the region and the red one denote the target gene; E, Sequence analysis revealed that the mutant had a single nucleotide transformation (A to G) at the end of the fifth intron.)
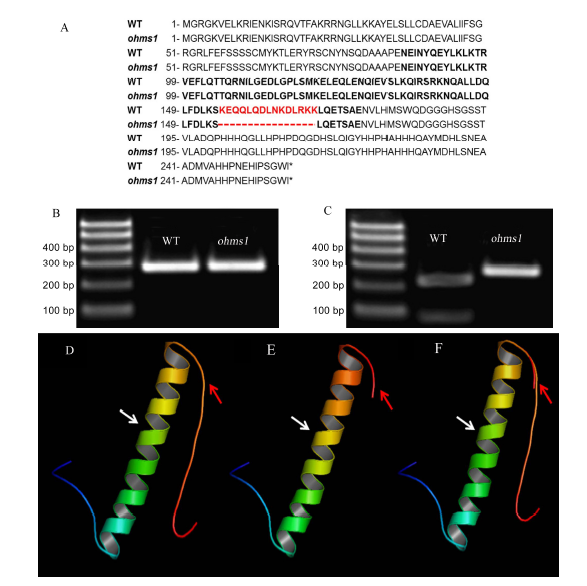
Fig. 3. Amino acid sequence analysis and protein structure of wild type (WT) and ohms1 mutant.(A, WT and ohms1 mutant protein sequences; The bold sequence are the OsMADS1 K-box domain and the red sequences denote the deletion of ohms1; B and C, Electrophoresis before and after Pst I enzyme digestion, respectively; D to F, Comparison of the 3D protein structure models of wild type and the mutant OsMADS1; D, Wild type structure; E, ohms1 mutant structure; F, Superposition of wild type and ohms1mutant sructures. The MADS domain and K-box domain of the OsMADS1 protein were represented by white arrows and red arrows, respectively.)
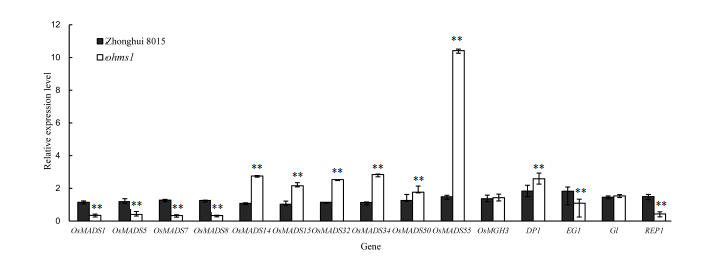
Fig. 4. qRT-PCR derived transcription profiles of a panel of genes associated with rice floral organ development. (** mean significant differences between wild type Zhonghui 8015 and ohms1 mutant at the 0.01 level. Bars represent the standard error.)

Fig. 5. Spikelet fertility of ohms1 mutant and F2 population.(A, Normal spikelet fertility of Zhobnghui 8015; B, Open hull and sterile spikelet of ohms1 mutant; C, Open hull but fertile spikelets in F2 population of ohms1/02428 (open hull but fertile spikelets are marked by red arrows).)
| [1] | Agrawal G K, Abe K, Yamazaki M, Miyao A, Hirochika H.2005. Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene.Plant Mol Biol, 59(1): 125-135. |
| [2] | Arora R, Agarwal P, Ray S, Singh A K, Singh V P, Tyagi A K, Kapoor S.2007. MADS-box gene family in rice genome-wide identification, organization and expression profiling during reproductive development and stress.BMC Genom, 8: 242-262. |
| [3] | Chen J J, Ding J H, Ouyang Y D, Du H Y, Yang J Y, Cheng K, Zhao J, Qiu S Q, Zhang X L, Yao J L, Liu K D, Wang L, Xu C G, Li X H, Xue Y B, Xia M, Ji Q, Lu J F, Xu M L, Zhang Q F.2008. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice.Proc Natl Acad Sci USA, 105(32): 11436-11441. |
| [4] | Chu H W, Qian Q, Liang W Q, Yin C S, Tan H X, Yao X, Yuan Z, Yang J, Huang H, Luo D, Ma H, Zhang D B.2006. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice.Plant Physiol, 142(3): 1039-1052. |
| [5] | Coen E S, Meyerowitz E M.1991. The war of the whorls: Genetic interactions controlling flower development.Nature, 353: 31-37. |
| [6] | Colombo L, Franken J, Koetje E, van Went J, Dons H J, Angenent G C, van Tunen A J.1995. The petunia MADS box gene FBP11 determines ovule identity.Plant Cell, 7(11): 1859-1868. |
| [7] | Cui R F, Han J K, Zhao S Z, Su K M, Wu F, Du X Q, Xu Q J, Chong K, Theissen G, Meng Z.2010. Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa).Plant J, 61(5): 767-781. |
| [8] | Diao Z J, Li S P, Duan Y L, Zheng L L, Zhou Y C, Wu W R.2013. Expression level of DDF1 affects vegetative growth and reproductive development in rice.Chin J Rice Sci, 27(4): 353-358. (in Chinese with English abstract) |
| [9] | Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky M F.2004. The SEP4 gene of Arahidopsis thaliana functions in floral organ and meristem identity.Curr Biol, 14(21): 1935-1940. |
| [10] | Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk P B, An G, Colombo L, Kater M M.2007. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice.Plant J, 52(4): 690-699. |
| [11] | Dreni L, Pilatone A, Yun D P, Erreni S, Pajoro A, Caporali E, Zhang D B, Kater M M.2011. Functional analysis of all agamous subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy.Plant Cell, 23(8): 2850-2863. |
| [12] | Ferrario S, Immink R G H, Shchennikova A, Busscher-Lange J, Angenent G C.2003. The MADS box gene FBP2 is required for SEPALLATA function in petunia.Plant Cell, 15(4): 914-925. |
| [13] | Fornara F, Parenicova L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura M M, Colombo L, Kater M M.2004. Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes.Plant Physiol, 135(4): 2207-2219. |
| [14] | Jang S, Lee B, Kim C, Kim S J, Yim J, Han J J, Lee S, Kim S R, An G.2003. The OsFOR1 gene encodes a polygalacturonase- inhibiting protein (PGIP) that regulates floral organ number in rice.Plant Mol Biol, 53(3): 357-372. |
| [15] | Jeon J S, Lee S, Jung K H, Yang W S, Yi G H, Oh B G, An G.2000a. Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes.Mol Breeding, 6(6): 581-592. |
| [16] | Jeon J S, Jang S, Lee S, Nam J, Kim C, Lee S H, Chung Y Y, Kim S R, Lee Y H, Cho Y G, An G.2000b. Leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development.Plant Cell, 12(6): 871-884. |
| [17] | Khanday I, Yadav S R, Vijayraghavan U.2013. Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways.Plant Physiol, 161(4): 1970-1983. |
| [18] | Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J.2012. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell, 24(5): 1848-1859. |
| [19] | Lamb R S, Irish V F.2003. Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages.Proc Natl Acad Sci USA, 100(11): 6558-6563. |
| [20] | Li Y F.2008. Map-based Cloning and Functional Analysis of LEAFY HULL STERILE 1-3 (lhs1-3) and PISTILLOID-STAMEN (PS) Genes in Rice (Oryza sativa L. ssp. indica). Chongqing, China: Southwest University. (in Chinese with English abstract) |
| [21] | Lohmann J U, Weigel D.2002. Building beauty: The genetic control of floral patterning.Dev Cell, 2(2): 135-142. |
| [22] | Lu Y J, Zheng K L.1992. A simple method for isolation of rice DNA.Chin J Rice Sci, 6(1): 47-48. (in Chinese with English abstract) |
| [23] | Lu S J, Wei H, Wang Y, Wang H M, Yang R F, Zhang X B, Tu J M.2012. Overexpression of a transcription factor OsMADS15 modifies plant architecture and flowering time in rice (Oryza sativa L.).Plant Mol Biol Rep, 30(6): 1461-1469. |
| [24] | Mandel M A, Yanofsky M F.1995. The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1.Plant Cell, 7(11): 1763-1771. |
| [25] | Moon S, Jung K H, Lee D E, Lee D Y, Lee J, An K, Kang H G, An G.2006. The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size.Mol Cells, 21(1): 147-152. |
| [26] | Nagasawa N, MiyOshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y.2003. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice.Development, 130(4): 705-718. |
| [27] | Prasad K, Parameswaran S, Vijayraghavan U.2005. OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs.Plant J, 43(6): 915-928. |
| [28] | Ren D Y, Li Y F, Zhao F M, Sang X C, Shi J Q, Wang N, Guo S, Ling Y H, Zhang C W, Yang Z L, He G H.2013. MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice.Plant Physiol, 162(2): 872-884. |
| [29] | Schmittgen T D, Livak K J.2008. Analyzing real-time PCR data by the comparative C(T) method.Nat Protoc, 3(6): 1101-1108. |
| [30] | Sentoku N, Kato H, Kitano H, Imai R.2005. OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy.Mol Genet Genom, 273(1): 1-9. |
| [31] | Suzaki T, Ohneda M, Toriba T, Yoshida A, Hirano H Y.2009. FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice.PLoS Genet, 5(10): 1-9. |
| [32] | Wang K J, Tang D, Hong L L, Xu W Y, Huang J, Li M, Gu M H, Xue Y B, Cheng Z K.2010. DEP and AFO regulate reproductive habit in rice.PLoS Genet, 6(1): 1-9. |
| [33] | Xiao H, Wang Y, Liu D F, Wang W M, Li X B, Zhao X F, Xu J C, Zhai W X, Zhu L H.2003. Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference.Plant Mol Biol, 52(5): 957-966. |
| [34] | Xiao H, Tang J F, Li Y F, Wang W M, Li X B, Jin L, Xie R, Luo H F, Zhao X F, Meng Z, He G H, Zhu L H.2009. STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice.Plant J, 59(5): 789-801. |
| [35] | Yadav S R, Prasad K, Vijayraghavan U.2007. Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ.Genetics, 176(1): 283-294. |
| [36] | Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano H Y.2004. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa.Plant Cell, 16(2): 500-509. |
| [37] | Yamaguchi T, Lee D Y, Miyao A, Hirochika H, An G, Hirano H Y.2006. Functional diversification of the two C-class MADS box genes OsMADS3 and OsMADS58 in Oryza sativa.Plant Cell, 18(1): 15-28. |
| [38] | Yang J Y, Zhao X B, Cheng K, Du H Y, Ouyang Y D, Chen J J, Qiu S Q, Huang J Y, Jiang Y H, Jiang L W, Ding J H, Wang J, Xu C G, Li X H, Zhang Q F.2012. A killer-protector system regulates both hybrid sterility and segregation distortion in rice.Science, 337: 1336-1340. |
| [39] | Yin L L, Xue H W.2012. The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development.Plant Cell, 24(3): 1049-1065. |
| [40] | Yoshida A, Suzaki T, Tanaka W, Hirano H Y.2009. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet.Proc Natl Acad Sci USA, 106: 20103-20108. |
| [41] | Zhang G L, Zhang S T, Xiao L T, Tang W B, Xiao Y H, Chen L Y.2014. Effect of high temperature stress on physiological characteristics of anther, pollen and stigma of rice during heading-flowering stage.Chin J Rice Sci, 28(2): 155-166. (in Chinese with English abstract) |
| [42] | Zhang Q F, Shen B Z, Dai X K, Mei M H, Saghai-Marool M A, Li Z B.1994. Using bulked extremes and ressessive classes to map genes for photoperiod-sensitive genic male sterility in rice.Proc Natl Acad Sci USA, 91(18): 8675-8679. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [13] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| [14] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| [15] | Zhang Guomei, Li Han, Liu Shanshan, Zhou Xuming, Lu Mingyang, Tang Liang, Sun Lihua. Water Extract of Rice False Smut Balls Activates Nrf2/HO-1 and Apoptosis Pathways, Causing Liver Injury [J]. Rice Science, 2023, 30(5): 473-485. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||