
Rice Science ›› 2022, Vol. 29 ›› Issue (1): 55-66.DOI: 10.1016/j.rsci.2021.12.005
• Research Paper • Previous Articles Next Articles
Tan Quanya#, Zhu Haitao#, Liu Hui, Ni Yuerong, Wu Shengze, Luan Xin, Liu Junwei, Yang Weifeng, Yang Zifeng, Zeng Ruizhen, Liu Guifu, Wang Shaokui( ), Zhang Guiquan(
), Zhang Guiquan( )
)
Received:2021-01-05
Accepted:2021-05-14
Online:2022-01-28
Published:2022-01-01
Contact:
Wang Shaokui, Zhang Guiquan
About author:First author contact:#These authors contributed equally to this work
Tan Quanya, Zhu Haitao, Liu Hui, Ni Yuerong, Wu Shengze, Luan Xin, Liu Junwei, Yang Weifeng, Yang Zifeng, Zeng Ruizhen, Liu Guifu, Wang Shaokui, Zhang Guiquan. Fine Mapping of QTLs for Stigma Exsertion Rate from Oryza glaberrima by Chromosome Segment Substitution[J]. Rice Science, 2022, 29(1): 55-66.
Add to citation manager EndNote|Ris|BibTeX
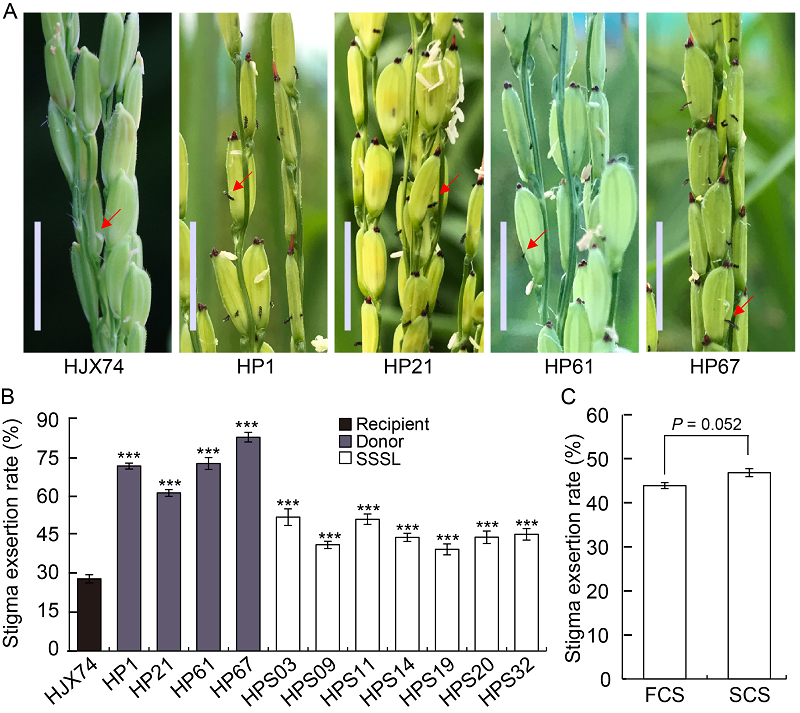
Fig. 1. Stigma exsertion rate (SER) of single-segment substitution lines (SSSLs) and their parents. A, Exserted stigmas in the panicles of parents. Red arrows point the exserted stigmas. Scale bars are 1 cm. B, Stigma exsertion rates of SSSLs and their donors. Huajingxian 74 (HJX74) was used as a control. ***, P ≤ ?0.001. C, Comparison of SER between the first cropping season (FCS) and the second cropping season (SCS) in seven SSSLs. Data in B and C are Mean ± SE (n = 3).
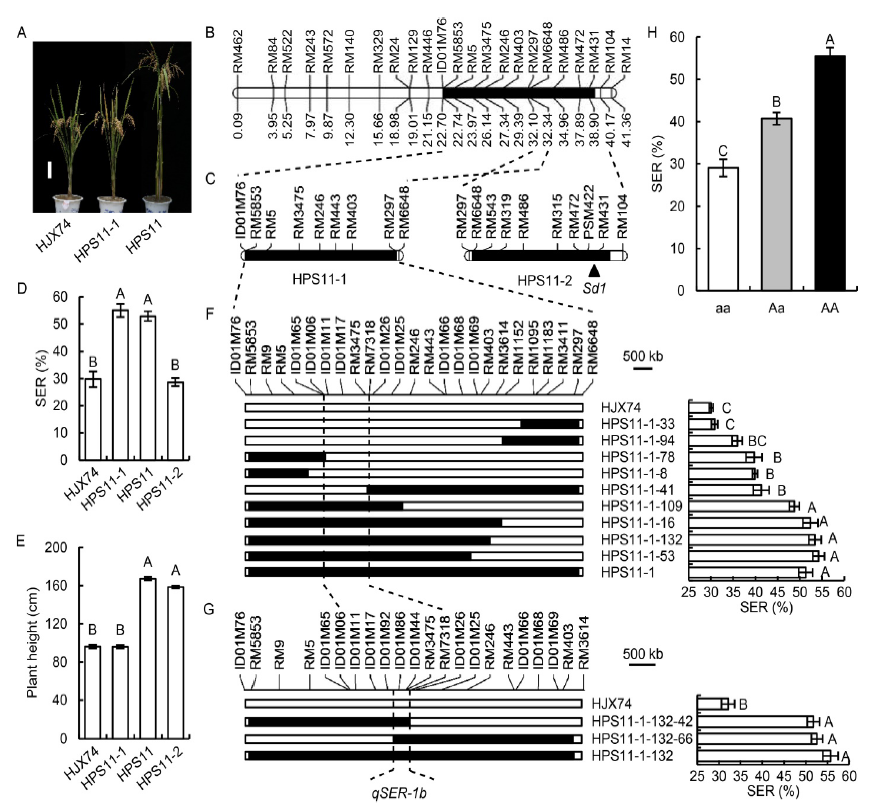
Fig. 2. Secondary substitution mapping of qSER-1b. A, Plant type of Huajingxian 74 (HJX74), HPS11 and HPS11-1. Scale bar is 15 cm. B, Substitution segment on chromosome 1 in HPS11. Physical distance (Mb) is shown under the chromosome. C, Substitution segments of HPS11-1 and HPS11-2. The position of Sd1 is pointed by the triangle (Sasaki et al, 2002). D, Stigma exsertion rate (SER) in HJX74, HPS11, HPS11-1 and HPS11-2. E, Plant height of HJX74, HPS11, HPS11-1 and HPS11-2. F, Secondary substitution mapping of qSER-1b based on the substitution segment of HPS11-1. G, Secondary substitution mapping of qSER-1b based on the substitution segment of HPS11-1-132. H, SER effects of three genotypes of qSER-1b in an F2 population. aa, Homozygous genotype of HJX74; Aa, Heterozygous genotype of HPS11-1/HJX74; AA, Homozygous genotype of HPS11-1. White and black blocks on chromosomes represent the genotypes of HJX74 and donor, respectively. Data in D-H are Mean ± SE (n = 3). Different uppercase letters above the bars indicate significant difference at the 0.001 level.
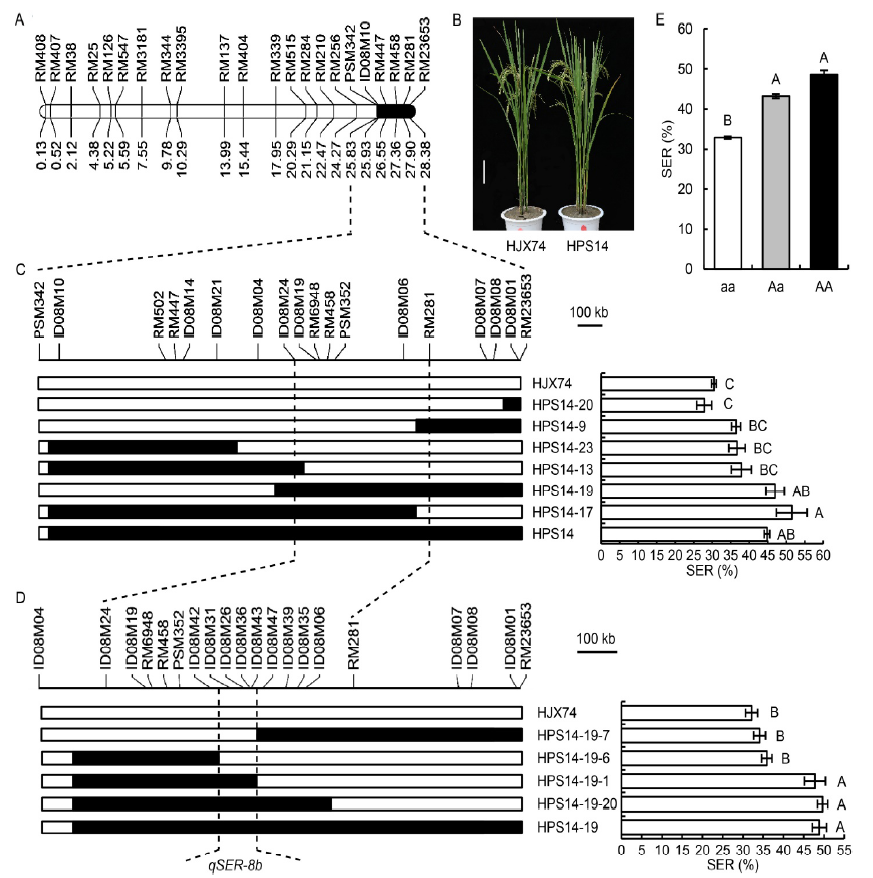
Fig. 3. Secondary substitution mapping of qSER-8b. A, Substitution segment on chromosome 8 in HPS14. Physical distance (Mb) is shown under the chromosome. B, Plant type of Huajingxian 74 (HJX74) and HPS14. Scale bar, 15 cm. C, Secondary substitution mapping based on the substitution segment of HPS14. D, Secondary substitution mapping of qSER-8b based on the substitution segment of HPS14-19. E, Stigma exsertion rate (SER) effects of three genotypes of qSER-8b in an F2 population. aa, Homozygous genotype of HJX74; Aa, Heterozygous genotype of HPS14/HJX74; AA, Homozygous genotype of HPS14. Data in C-E are Mean ± SE (n = 3). Different uppercase letters above bars indicate significant difference at the 0.001 level.
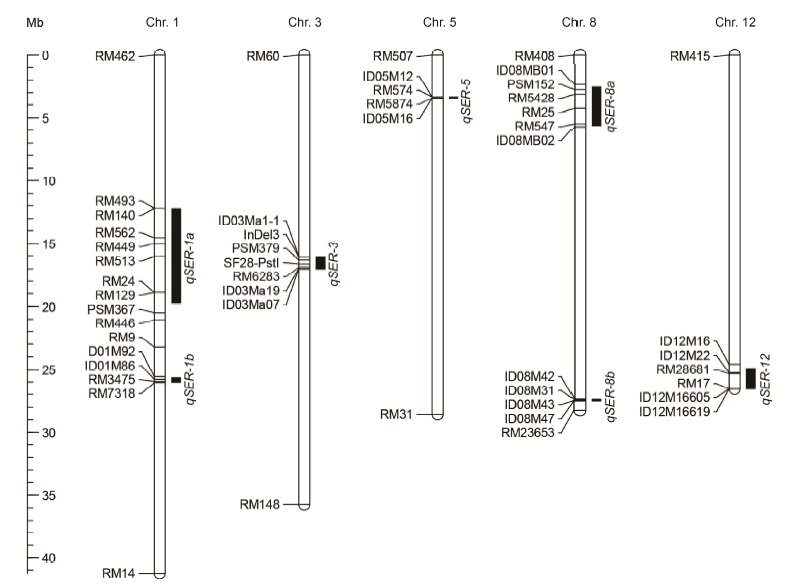
Fig. 4. Chromosomal locations of the seven QTLs of SER in single-segment substitution lines. Black bars on the right of each chromosome were the estimated intervals of QTLs with their names on the right. SER, Stigma exsertion rate; Chr., Chromosome; Mb, Megabase.
| QTL | Chromosome | Interval (kb) | Estimated length (kb) | Maximum length (kb) | Additive effect (%) |
|---|---|---|---|---|---|
| qSER-1a | 1 | 12 291.6-19 797.0 | 7 505.4 | 8 302.9 | 8.0 ± 0.7 |
| qSER-1b | 1 | 25 759.6-26 092.6 | 333.0 | 500.2 | 11.5 ± 1.0 |
| qSER-3 | 3 | 16 161.4-17 105.0 | 943.5 | 965.1 | 11.9 ± 1.1 |
| qSER-5 | 5 | 3 448.9-3 541.4 | 92.5 | 104.0 | 8.6 ± 0.6 |
| qSER-8a | 8 | 2 685.9-5 716.0 | 3 030.1 | 3 360.3 | 5.7 ± 0.5 |
| qSER-8b | 8 | 27 510.3-27 617.8 | 107.5 | 152.5 | 8.0 ± 0.7 |
| qSER-12 | 12 | 25 351.7-26 992.7 | 1 641.0 | 1 980.5 | 6.5 ± 0.6 |
Table 1. Additive effects of QTLs for stigma exsertion rate (SER) detected in single-segment substitution lines.
| QTL | Chromosome | Interval (kb) | Estimated length (kb) | Maximum length (kb) | Additive effect (%) |
|---|---|---|---|---|---|
| qSER-1a | 1 | 12 291.6-19 797.0 | 7 505.4 | 8 302.9 | 8.0 ± 0.7 |
| qSER-1b | 1 | 25 759.6-26 092.6 | 333.0 | 500.2 | 11.5 ± 1.0 |
| qSER-3 | 3 | 16 161.4-17 105.0 | 943.5 | 965.1 | 11.9 ± 1.1 |
| qSER-5 | 5 | 3 448.9-3 541.4 | 92.5 | 104.0 | 8.6 ± 0.6 |
| qSER-8a | 8 | 2 685.9-5 716.0 | 3 030.1 | 3 360.3 | 5.7 ± 0.5 |
| qSER-8b | 8 | 27 510.3-27 617.8 | 107.5 | 152.5 | 8.0 ± 0.7 |
| qSER-12 | 12 | 25 351.7-26 992.7 | 1 641.0 | 1 980.5 | 6.5 ± 0.6 |
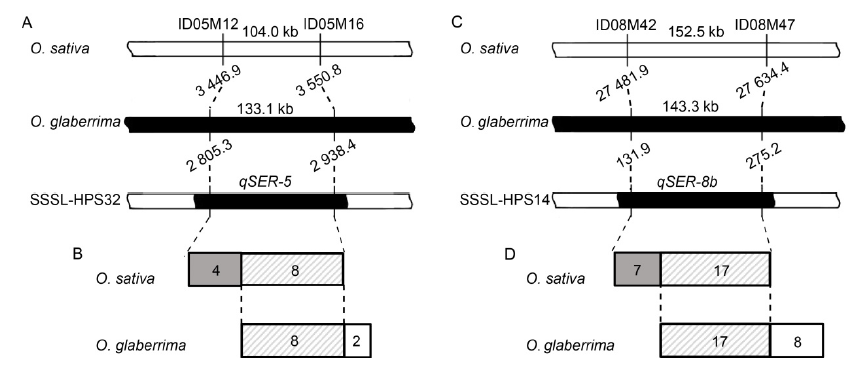
Fig. 5. Maximum intervals and numbers of open reading frames (ORFs) of qSER-5 and qSER-8b in genomes of O. sativa and O. glaberrima. A, Maximum interval of qSER-5 on chromosome 5 of the two genomes. B, Number of ORFs of the qSER-5 interval identified in the two genomes. C, Maximum interval of qSER-8b on chromosome 8 of the two genomes. D, Number of ORFs of the qSER-8b interval identified in the two genomes. In A and C, white and black blocks represent respectively the chromosome segments of O. sativa and O. glaberrima. In B and D, the number in the shaded box is the number of ORFs identified in the O. sativa and O. glaberrima genomes, whereas the number in the gray box is the number of ORFs identified only in O. sativa genome, and the number in white box is the number of ORFs identified only in O. glaberrima genome.
| [1] | Bakti C, Tanaka J. 2019. Detection of dominant QTLs for stigma exsertion ratio in rice derived from Oryza rufipogon accession ‘W0120’. Breed Sci, 69(1): 143-150. |
| [2] | Cai J, Liao Q P, Dai Z J, Zhu H T, Zeng R Z, Zhang Z M, Zhang G Q. 2013. Allelic differentiations and effects of the Rf3 and Rf4 genes on fertility restoration in rice with wild abortive cytoplasmic male sterility. Biol Plant, 57(2): 274-280. |
| [3] | Dai Z J, Lu Q, Luan X, Cai J, Zhu H T, Liu Z Q, Zeng R Z, Zhang Z M, Wang S K, Zheng L J, Li J L, Zhang G Q. 2015. Development of a platform for breeding by design of CMS lines based on an SSSL library in rice (Oryza sativa L.). Euphytica, 205(1): 63-72. |
| [4] | Dai Z J, Lu Q, Luan X, Ouyang L, Guo J, Liang J Y, Zhu H T, Wang W J, Wang S K, Zeng R Z, Liu Z Q, Zhang Z M, Zhu X Y, Zhang G Q. 2016. Development of a platform for breeding by design of CMS restorer lines based on an SSSL library in rice (Oryza sativa L.). Breed Sci, 66(5): 768-775. |
| [5] | Dang X J, Liu E B, Liang Y F, Liu Q M, Breria C M, Hong D L. 2016. QTL detection and elite alleles mining for stigma traits in Oryza sativa by association mapping. Front Plant Sci, 7: 1188. |
| [6] | Deng Y D, Ying J Z, Shi Y Y, Xiao C L, Zhang H Q. 2010. Mapping of QTLs for percentage of exserted stigma in rice. J Hunan Agric Univ, 36(4): 373-376. (in Chinese with English abstract) |
| [7] |
Eshed Y, Zamir D. 1995. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics, 141(3): 1147-1162.
PMID |
| [8] | Fang C W, Li L, He R M, Wang D Q, Wang M, Hu Q, Ma Q R, Qin K Y, Feng X Y, Zhang G Q, Fu X L, Liu Z Q. 2019. Identification of S23causing both interspecific hybrid male sterility and environment-conditioned male sterility in rice. Rice, 12(1): 10. |
| [9] | Feng L L, Jing Y H, Huang C, Xu Z J, Chen W F. 2010. QTL analysis of percentage of exserted stigma in rice (Oryza sativa L.). North Rice, 40(3): 20-22. (in Chinese with English abstract) |
| [10] | Guo L, Qiu F L, Gandhi H, Kadaru S, De Asis E J, Zhuang J Y, Xie F M. 2017. Genome-wide association study of outcrossing in cytoplasmic male sterile lines of rice. Sci Rep, 7(1): 3223. |
| [11] | He N, Wu R X, Pan X P, Peng L P, Sun K, Zou T, Zhu H T, Zeng R Z, Liu Z Q, Liu G F, Wang S K, Zhang G Q, Fu X L. 2017. Development and trait evaluation of chromosome single- segment substitution lines of O. meridionalis in the background of O. sativa. Euphytica, 213: 281. |
| [12] | Hu S P, Zhou Y, Zhang L, Zhu X D, Wang Z G, Li L, Luo L J, Zhou Q M. 2009. QTL analysis of floral traits of rice (Oryza sativa L.) under well-watered and drought stress conditions. Genes Genom, 31(2): 173-181. |
| [13] | Huang X H, Kurata N, Wei X H, Wang Z X, Wang A H, Zhao Q, Zhao Y, Liu K Y, Lu H Y, Li W J, Guo Y L, Lu Y Q, Zhou C C, Fan D L, Weng Q J, Zhu C R, Huang T, Zhang L, Wang Y C, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan X P, Xu Q, Dong G J, Zhan Q L, Li C Y, Fujiyama A, Toyoda A, Lu T T, Feng Q, Qian Q, Li J Y, Han B. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature, 490: 497-501. |
| [14] | Jiang J H, Xu L, Xiao M H, Hu C M, Zhang Y, Wang D Z, Dang X J. 2021. Genetic analysis and QTLs identification of stigma traits in japonica rice (Oryza sativa L.). Euphytica, 217: 82. |
| [15] | Kato H, Namai H. 1987. Floral characteristics and environmental factors for increasing natural outcrossing rate for F1 hybrid seed production of rice Oryza sativa L. Jpn J Breed, 37(3): 318-330. |
| [16] | Lalitha S. 2000. Primer premier 5. Biotech Softw Internet Rep, 1(6): 270-272. |
| [17] | Li C, Sun C Q, Mu P, Chen L, Wang X K. 2001. QTL analysis of anther length and ratio of stigma exsertion, two key traits of classification for cultivated rice (Oryza sativa L.) and common wild rice (O. rufipogon Griff.). Acta Genet Sin, 28(8): 746-751. (in Chinese with English abstract) |
| [18] | Li F P, Gao Y H, Wu B Q, Cai Q P, Zhan P L, Yang W F, Shi W X, Li X H, Yang Z F, Tan Q Y, Luan X, Zhang G Q, Wang S K. 2021. High-quality de novo genome assembly of Huajingxian 74, a receptor parent of single segment substitution lines. Rice Sci, 28(2): 109-113. |
| [19] | Li H B, Gao F Y, Zeng L H, Li Q X, Lu X J, Li Z H, Ren J S, Su X W, Ren G J. 2010. QTL analysis of rice stigma morphology using an introgression line from Oryza longistaminata L. Mol Plant Breed, 8(6): 1082-1089. (in Chinese with English abstract) |
| [20] | Li P B, Feng F C, Zhang Q L, Chao Y, Gao G J, He Y Q. 2014a. Genetic mapping and validation of quantitative trait loci for stigma exsertion rate in rice. Mol Breed, 34: 2131-2138. |
| [21] | Li P B, Su G C, Feng F C, Wang P, Yu S B, He Y Q. 2014b. Mapping of minor quantitative trait loci (QTLs) conferring fertility restoration of wild abortive cytoplasmic male sterility and QTLs conferring stigma exsertion in rice. Plant Breed, 133(6): 722-727. |
| [22] | Li T, Chen Y W. 1985. Genetics of stigma exsertion in rice. Rice Genet Newsl, 2: 84-85. |
| [23] | Li W, Sheng Z H, Zhu Z L, Wei X J, Shi L, Wu Y W, Tang S Q, Wang J L, Hu P S. 2017. QTL mapping of japonica rice stigma exsertion rate. Chin J Rice Sci, 31(1): 23-30. (in Chinese with English abstract) |
| [24] | Li W H, Dong G J, Hu X M, Teng S, Guo L B, Zeng D L, Qiao Q. 2003. QTL analysis for percentage of exserted stigma in rice (Oryza sativa L). Acta Genet Sin, 30(7): 637-640. (in Chinese with English abstract) |
| [25] |
Liu Y, Zhang A N, Wang F M, Kong D Y, Li M S, Bi J G, Zhang F Y, Wang J H, Luo X X, Pan Z Q, Yu X Q, Liu G L, Luo L J. 2019. Fine mapping a quantitative trait locus, qSER-7, that controls stigma exsertion rate in rice (Oryza sativa L.). Rice, 12(1): 46.
PMID |
| [26] | Lou J, Yue G H, Yang W Q, Mei H W, Luo L J, Lu H J. 2014. Mapping QTLs influencing stigma exertion in rice. Bulg J Agric Sci, 20(6): 1450-1456. |
| [27] | Luan X, Dai Z J, Yang W F, Tan Q Y, Lu Q, Guo J, Zhu H T, Liu G F, Wang S K, Zhang G Q. 2019. Breeding by design of CMS lines on the platform of SSSL library in rice. Mol Breed, 39: 126. |
| [28] | Ma X, Zheng Z, Lin F S, Ge T T, Sun H M. 2018. Genetic analysis and gene mapping of a low stigma exposed mutant gene by high-throughput sequencing. PLoS One, 13(1): e0186942. |
| [29] |
Mackay T F C. 2014. Epistasis and quantitative traits: Using model organisms to study gene-gene interactions. Nat Rev Genet, 15(1): 22-33.
PMID |
| [30] | Mahalingam A, Saraswathi R, Ramalingam J, Jayaraj T. 2013. Genetics of floral traits in Cytoplasmic male sterile (CMS) and restorer lines of hybrid rice (Oryza sativa L.). Pak J Bot, 45(6): 1897-1904. |
| [31] | Marathi B, Jena K K. 2015. Floral traits to enhance outcrossing for higher hybrid seed production in rice: Present status and future prospects. Euphytica, 201: 1-14. |
| [32] | Marathi B, Ramos J, Hechanova S L, Oane R H, Jena K K. 2015. SNP genotyping and characterization of pistil traits revealing a distinct phylogenetic relationship among the species of Oryza. Euphytica, 201: 131-148. |
| [33] | McCouch S R, Cho Y G, Yano M, Paul E, Blinstrub M, Morishima H, Kinosita T. 1997. Report on QTL nomenclature. Rice Genet Newsl, 14: 11-12. |
| [34] |
Miyata M, Yamamoto T, Komori T, Nitta N. 2007. Marker- assisted selection and evaluation of the QTL for stigma exsertion under japonica rice genetic background. Theor Appl Genet, 114: 539-548.
PMID |
| [35] |
Murray M G, Thompson W F. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res, 8(19): 4321-4325.
PMID |
| [36] | National Research Council. 1996. Lost crops of Africa:Grains. Washington, USA: National Academies Press: 17. |
| [37] | Oka H I. 1958. Intervarietal variation and classification of cultivated rice. Ind J Genet Plant Breed, 18: 78-89. |
| [38] | Parmar K S, Siddiq E A, Swaminathan M S. 1979. Variation in components of flowering behavior of rice. Ind J Genet Plant Breed, 39(3): 542-550. |
| [39] | Qian Q, Guo L B, Smith S M, Li J Y. 2016. Breeding high-yield superior quality hybrid super rice by rational design. Natl Sci Rev, 3(3): 283-294. |
| [40] | Rahman M H, Yu P, Zhang Y X, Sun L P, Wu W X, Shen X H, Zhan X D, Chen D B, Cao L Y, Cheng S H. 2016. Quantitative trait loci mapping of the stigma exertion rate and spikelet number per panicle in rice (Oryza sativa L.). Genet Mol Res, 15(4): gmr15048432. |
| [41] | Rahman M H, Zhang Y X, Sun L P, Zhang K Q, Rahman M S, Wu W X, Zhan X D, Cao L Y, Cheng S H. 2017a. Genetic mapping of quantitative trait loci for the stigma exsertion rate in rice (Oryza sativa L.). J Integr Agric, 16(7): 1423-1431. |
| [42] | Rahman M H, Zhang Y X, Zhang K Q, Rahman M S, Barman H N, Riaz A, Chen Y Y, Wu W X, Zhan X D, Cao L Y, Cheng S H. 2017b. Genetic dissection of the major quantitative trait locus (qSE11), and its validation as the major influence on the rate of stigma exsertion in rice (Oryza sativa L.). Front Plant Sci, 81818. |
| [43] | Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush G S, Kitano H, Matsuoka M. 2002. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature, 416: 701-702. |
| [44] | Sui F Q, Zhao D K, Zhu H T, Gong Y F, Tang Z, Huang X Y, Zhang G Q, Zhao F J. 2019. Map-based cloning of a new total loss-of-function allele of OsHMA3 causes high cadmium accumulation in rice grain. J Exp Bot, 70(10): 2857-2871. |
| [45] | Takano-Kai N, Doi K, Yoshimura A. 2011. GS3 participates in stigma exsertion as well as seed length in rice. Breed Sci, 61: 244-250. |
| [46] | Tan Q Y, Zou T, Zheng M M, Ni Y R, Luan X, Li X H, Yang W F, Yang Z F, Zhu H T, Zeng R Z, Liu G F, Wang S K, Fu X L, Zhang G Q. 2020. Substitution mapping of the major quantitative trait loci controlling stigma exsertion rate from Oryza glumaepatula. Rice, 13(1): 37. |
| [47] | Tan Q Y, Wang C S, Luan X, Zheng L J, Ni Y R, Yang W F, Yang Z F, Zhu H T, Zeng R Z, Liu G F, Wang S K, Zhang G Q. 2021. Dissection of closely linked QTLs controlling stigma exsertion rate in rice by substitution mapping. Theor Appl Genet, 134(4): 1253-1262. |
| [48] | Teng B, Zeng R Z, Wang Y C, Liu Z Q, Zhang Z M, Zhu H T, Ding X H, Li W T, Zhang G Q. 2012. Detection of allelic variation at the Wx locus with single-segment substitution lines in rice (Oryza sativa L.). Mol Breed, 30: 583-595. |
| [49] |
Uga Y, Fukuta Y, Cai H W, Iwata H, Ohsawa R, Morishima H, Fujimura T. 2003. Mapping QTLs influencing rice floral morphology using recombinant inbred lines derived from a cross between Oryza sativa L. and Oryza rufipogon Griff. Theor Appl Genet, 107: 218-226.
PMID |
| [50] | Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D. 2012. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet, 44(8): 950-954. |
| [51] | Wu C, Cui K H, Hu Q Q, Wang W C, Nie L X, Huang J L, Peng S B. 2019. Enclosed stigma contributes to higher spikelet fertility for rice (Oryza sativa L.) subjected to heat stress. Crop J, 7(3): 335-349. |
| [52] |
Xi Z Y, He F H, Zeng R Z, Zhang Z M, Ding X H, Li W T, Zhang G Q. 2006. Development of a wide population of chromosome single-segment substitution lines in the genetic background of an elite cultivar of rice (Oryza sativa L.). Genome, 49(5): 476-484.
PMID |
| [53] | Xie Y Y, Xu P, Huang J L, Ma S J, Xie X R, Tao D Y, Chen L T, Liu Y G. 2017. Interspecific hybrid sterility in rice is mediated by OgTPR1 at the S1 locus encoding a peptidase-like protein. Mol Plant, 10(8): 1137-1140. |
| [54] | Xiong L Z, Liu K D, Dai X K, Xu C G, Zhang Q. 1999. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor Appl Genet, 98(2): 243-251. |
| [55] | Xu S L, Zheng Y C, Liu Y, Guo X H, Tan Y Y, Qian Q P, Shu Q Y, Huang J Z. 2019. Identification of a major quantitative trait locus and its candidate underlying genetic variation for rice stigma exsertion rate. Crop J, 7(3): 350-359. |
| [56] | Yamamoto T, Takemori N, Sue N. 2003. QTL analysis of stigma exsertion in rice. Rice Genet Newsl, 10: 33-34. |
| [57] | Yan W G, Li Y, Agrama H A, Luo D G, Gao F Y, Lu X J, Ren G J. 2009. Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.). Mol Breed, 24: 277-292. |
| [58] | Yang W F, Liang J Y, Hao Q W, Luan X, Tan Q Y, Lin S W, Zhu H T, Liu G F, Liu Z P, Bu S H, Wang S K, Zhang G Q. 2021. Fine mapping of two grain chalkiness QTLs sensitive to high temperature in rice. Rice, 14(1): 33. |
| [59] | Yang W N, Guo Z L, Huang C L, Duan L F, Chen G X, Jiang N, Fang W, Feng H, Xie W B, Lian X M, Wang G W, Luo Q M, Zhang Q F, Liu Q, Xiong L Z. 2014. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun, 5: 5087. |
| [60] | Yue G H, Mei H W, Pan B R, Lou J, Li M S, Luo L J. 2009. Mapping of QTLs affecting stigma exsertion rate of Huhan 1B as a CMS maintainer of upland hybrid rice. Acta Agric Zhejiangensis, 21(3): 241-245. (in Chinese with English abstract) |
| [61] | Zhang G Q. 2019. The platform of breeding by design based on the SSSL library in rice. Hereditas, 41(8): 754-760. (in Chinese with English abstract) |
| [62] | Zhang G Q, Zeng R Z, Zhang Z M, Ding X H, Li W T, Liu G M, He F H, Tulukdar A, Huang C F, Xi Z Y, Qin L J, Shi J Q, Zhao F M, Feng M J, Shan Z L, Chen L, Guo X Q, Zhu H T, Lu Y G. 2004. The construction of a library of single segment substitution lines in rice (Oryza sativa L.). Rice Genet Newsl, 21 : 85-87. |
| [63] | Zhang K Q, Zhang Y X, Wu W X, Zhan X D, Anis G B, Rahman M H, Hong Y B, Riaz A, Zhu A K, Cao Y R, Sun L P, Yang Z F, Yang Q Q, Cao L Y, Cheng S H. 2018. qSE7 is a major quantitative trait locus (QTL) influencing stigma exsertion rate in rice (Oryza sativa L.). Sci Rep, 8: 14523. |
| [64] | Zhao F M, Zhu H T, Zeng R Z, Zhang G Q, Xu S Z. 2016. Detection of additive and additive × environment interaction effects of QTLs for yield-component traits of rice using single- segment substitution lines (SSSLs). Plant Breed, 135(4): 452-458. |
| [65] | Zhao H W, Sun L L, Xiong T Y, Wang Z Q, Liao Y, Zou T, Zheng M M, Zhang Z, Pan X P, He N, Zhang G Q, Zhu H T, Liu Z Q, He P, Fu X L. 2019. Genetic characterization of the chromosome single-segment substitution lines of O. glumaepatula and O. barthii and identification of QTLs for yield-related traits. Mol Breed, 39: 51. |
| [66] |
Zhou H, Li P B, Xie W B, Hussain S, Li Y B, Xia D, Zhao H, Sun S Y, Chen J X, Ye H, Hou J, Zhao D, Gao G J, Zhang Q L, Wang G W, Lian X M, Xiao J H, Yu S B, Li X H, He Y Q. 2017. Genome-wide association analyses reveal the genetic basis of stigma exsertion in rice. Mol Plant, 10(4): 634-644.
PMID |
| [67] | Zhou Y L, Xie Y H, Cai J L, Liu C B, Zhu H T, Jiang R, Zhong Y Y, Zhang G L, Tan B, Liu G F, Fu X L, Liu Z Q, Wang S K, Zhang G Q, Zeng R Z. 2017. Substitution mapping of QTLs controlling seed dormancy using single segment substitution lines derived from multiple cultivated rice donors in seven cropping seasons. Theor Appl Genet, 130(6): 1191-1205. |
| [68] | Zou T, Zhao H W, Li X H, Zheng M M, Zhang S D, Sun L L, He N, Pan X P, Liu Z Q, Fu X L. 2020. QTLs detection and pyramiding for stigma exsertion rate in wild rice species by using the single-segment substitution lines. Mol Breed, 40: 74. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [3] | Si Fengfeng, Fan Fengfeng, Wei Xiao, He Shihao, Li Xianlong, Peng Xiaojue, Li Shaoqing. Quantitative Trait Locus Mapping of High Photosynthetic Efficiency and Biomass in Oryza longistaminata [J]. Rice Science, 2022, 29(6): 569-576. |
| [4] | Kossi Lorimpo Adjah, Maxwell Darko Asante, Aboubacar Toure, Mawuli Aziadekey, Francis Osei Amoako-Andoh, Michael Frei, Yacouba Diallo, Komi Agboka. Improvement of Rice Production under Drought Conditions in West Africa: Application of QTLs in Breeding for Drought Resistance [J]. Rice Science, 2022, 29(6): 512-521. |
| [5] | Nie Yuanyuan, Xia Hui, Ma Xiaosong, Lou Qiaojun, Liu Yi, Zhang Anling, Cheng Liang, Yan Longan, Luo Lijun. Dissecting Genetic Basis of Deep Rooting in Dongxiang Wild Rice [J]. Rice Science, 2022, 29(3): 277-287. |
| [6] | Hui Wang, Jiayu Zhang, Farkhanda Naz, Juan Li, Shuangfei Sun, Guanghua He, Ting Zhang, Yinghua Ling, Fangming Zhao. Identification of Rice QTLs for Important Agronomic Traits with Long-Kernel CSSL-Z741 and Three SSSLs [J]. Rice Science, 2020, 27(5): 414-422. |
| [7] | Yaobin Qin, Peng Cheng, Yichen Cheng, Yue Feng, Derun Huang, Tingxu Huang, Xianjun Song, Jiezheng Ying. QTL-Seq Identified a Major QTL for Grain Length and Weight in Rice Using Near Isogenic F2 Population [J]. Rice Science, 2018, 25(3): 121-131. |
| [8] | Vivitha P., Raveendran M., Vijayalakshmi D.. Introgression of QTLs Controlling Spikelet Fertility Maintains Membrane Integrity and Grain Yield in Improved White Ponni Derived Progenies Exposed to Heat Stress [J]. Rice Science, 2017, 24(1): 32-40. |
| [9] | Chuan Tong, Lei Liu, L. E. Waters Daniel, Jin-song Bao. Association Mapping and Marker Development of Genes for Starch Lysophospholipid Synthesis in Rice [J]. Rice Science, 2016, 23(6): 287-296. |
| [10] | ZUO Shi-min, ZHANG Ya-fang, CHEN Zong-xiang, JIANG Wei, FENG Ming-hui, PAN Xue-biao. Improvement of Rice Resistance to Sheath Blight by Pyramiding QTLs Conditioning Disease Resistance and Tiller Angle [J]. RICE SCIENCE, 2014, 21(6): 318-326. |
| [11] | ZHANG Hong-jun, QU Li-jun, XIANG Chao, WANG Hui, XIA Jia-fa, LI Ze-fu, GAO Yong-ming, SHI Ying-yao. Dissection of Genetic Mechanism of Abnormal Heading in Hybrid Rice [J]. RICE SCIENCE, 2014, 21(4): 201-209. |
| [12] | LUO Li-li1, 2, #, ZHANG Ying-xin1, #, CHEN Dai-bo1, ZHAN Xiao-deng1, SHEN Xi-hong1, CHENG Shi-hua1, CAO Li-yong1. QTL Mapping for Hull Thickness and Related Traits in Hybrid Rice Xieyou 9308 [J]. RICE SCIENCE, 2014, 21(1): 29-38. |
| [13] | ZHANG Chang-quan, HU Bing, ZHU Kong-zhi, ZHANG Hua, LENG Ya-lin, TANG Shu-zhu, GU Ming-hong, LIU Qiao-quan. QTL Mapping for Rice RVA Properties Using High-Throughput Re-sequenced Chromosome Segment Substitution Lines [J]. RICE SCIENCE, 2013, 20(6): 407-414. |
| [14] | TIAN Fu-kuan1, 2, #, RUAN Ban-pu1, 3, #, YAN Mei-xian1, YE Shi-fang1, PENG You-lin1, DONG Guo-jun1, ZHU Li1, HU Jiang1, YAN Hong-lan1, GUO Long-biao1, QIAN Qian1, GAO Zhen-yu1. Genetic Analysis and QTL Mapping of Mature Seed Culturability in Indica Rice [J]. RICE SCIENCE, 2013, 20(5): 313-319. |
| [15] | HU Wen-de, ZHANG Hong, JIANG Jian-hua, WANG Ying-ying, SUN Da-yun, WANG Xiao-shuai, LIANG Kui, HONG De-lin. Genetic Analysis and QTL Mapping of Large Flag Leaf Angle Trait in Japonica Rice [J]. RICE SCIENCE, 2012, 19(4): 277-285. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||