
Rice Science ›› 2022, Vol. 29 ›› Issue (6): 522-534.DOI: 10.1016/j.rsci.2022.03.001
• Research Paper • Previous Articles Next Articles
Liu Yantong#, Li Ting#, Jiang Zhishu#, Zeng Chuihai, He Rong, Qiu Jiao, Lin Xiaoli, Peng Limei, Song Yongping, Zhou Dahu, Cai Yicong, Zhu Changlan, Fu Junru, He Haohua( ), Xu Jie(
), Xu Jie( )
)
Received:2022-01-27
Accepted:2022-03-28
Online:2022-11-28
Published:2022-09-09
Contact:
He Haohua, Xu Jie
About author:#These authors contributed equally to this work
Liu Yantong, Li Ting, Jiang Zhishu, Zeng Chuihai, He Rong, Qiu Jiao, Lin Xiaoli, Peng Limei, Song Yongping, Zhou Dahu, Cai Yicong, Zhu Changlan, Fu Junru, He Haohua, Xu Jie. Characterization of a Novel Weak Allele of RGA1/D1 and Its Potential Application in Rice Breeding[J]. Rice Science, 2022, 29(6): 522-534.
Add to citation manager EndNote|Ris|BibTeX

Fig. 1. Phenotypic characterization of XLX, HZ, CHT025 and their F1 plants. A and B, Phenotypes of XLX, HZ, CHT025 and their F1 plants at the filling stage. Scale bars, 20 cm. C, Grain shapes of XLX, HZ, CHT025 and their F1s. Scale bar, 5 mm. XLX, Xiaolixiang; HZ, Huazhan; CHT025, Changhui T025.
| Rice material | PH (cm) | EPN | PL (cm) | PBN | SBN | GNPS | SSR (%) | TGW (g) | YPP (g) |
|---|---|---|---|---|---|---|---|---|---|
| XLX | 112.1 ± 3.5 | 15.7 ± 2.7 | 23.5 ± 0.6 | 14.0 ± 0.4 | 53.6 ± 1.6 | 298.2 ± 19.6 | 90.6 ± 1.2 | 16.3 ± 0.1 | 70.1 ± 5.9 |
| TN1 | 108.1 ± 2.2 | 12.4 ± 1.7 | 25.9 ± 0.6 | 12.3 ± 0.4 | 24.5 ± 3.3 | 177.8 ± 16.7 | 83.3 ± 1.8 | 26.2 ± 0.6 | 48.0 ± 4.0 |
| CHT025 | 106.3 ± 1.9 | 9.1 ± 0.7 | 29.0 ± 1.2 | 13.0 ± 0.2 | 58.2 ± 5.4 | 386.0 ± 6.0 | 95.5 ± 0.7 | 17.6 ± 0.2 | 59.4 ± 4.1 |
| HZ | 110.5 ± 1.8 | 11.8 ± 0.7 | 24.1 ± 1.1 | 12.4 ± 0.3 | 26.5 ± 2.3 | 183.9 ± 20.2 | 87.3 ± 0.3 | 24.6 ± 0.8 | 46.8 ± 2.0 |
| XLX/TN1 F1 | 183.0 ± 3.8 | 20.2 ± 1.9 | 30.2 ± 0.5 | 13.8 ± 0.6 | 71.7 ± 3.2 | 346.3 ± 17.6 | 94.6 ± 0.7 | 21.2 ± 0.2 | 140.0 ± 7.1 |
| XLX/CHT025 F1 | 169.0 ± 2.3 | 16.0 ± 0.2 | 31.3 ± 0.4 | 13.7 ± 0.7 | 72.0 ± 5.1 | 344.8 ± 22.3 | 93.7 ± 0.5 | 18.0 ± 0.1 | 103.1 ± 5.3 |
| XLX/HZ F1 | 164.6 ± 1.5 | 18.0 ± 1.2 | 31.1 ± 0.4 | 14.4 ± 0.5 | 71.9 ± 4.5 | 334.3 ± 18.2 | 94.2 ± 0.1 | 21.0 ± 0.2 | 119.1 ± 6.4 |
Table 1. Agronomic characteristics of XLX and three common varieties as well as their F1 plants.
| Rice material | PH (cm) | EPN | PL (cm) | PBN | SBN | GNPS | SSR (%) | TGW (g) | YPP (g) |
|---|---|---|---|---|---|---|---|---|---|
| XLX | 112.1 ± 3.5 | 15.7 ± 2.7 | 23.5 ± 0.6 | 14.0 ± 0.4 | 53.6 ± 1.6 | 298.2 ± 19.6 | 90.6 ± 1.2 | 16.3 ± 0.1 | 70.1 ± 5.9 |
| TN1 | 108.1 ± 2.2 | 12.4 ± 1.7 | 25.9 ± 0.6 | 12.3 ± 0.4 | 24.5 ± 3.3 | 177.8 ± 16.7 | 83.3 ± 1.8 | 26.2 ± 0.6 | 48.0 ± 4.0 |
| CHT025 | 106.3 ± 1.9 | 9.1 ± 0.7 | 29.0 ± 1.2 | 13.0 ± 0.2 | 58.2 ± 5.4 | 386.0 ± 6.0 | 95.5 ± 0.7 | 17.6 ± 0.2 | 59.4 ± 4.1 |
| HZ | 110.5 ± 1.8 | 11.8 ± 0.7 | 24.1 ± 1.1 | 12.4 ± 0.3 | 26.5 ± 2.3 | 183.9 ± 20.2 | 87.3 ± 0.3 | 24.6 ± 0.8 | 46.8 ± 2.0 |
| XLX/TN1 F1 | 183.0 ± 3.8 | 20.2 ± 1.9 | 30.2 ± 0.5 | 13.8 ± 0.6 | 71.7 ± 3.2 | 346.3 ± 17.6 | 94.6 ± 0.7 | 21.2 ± 0.2 | 140.0 ± 7.1 |
| XLX/CHT025 F1 | 169.0 ± 2.3 | 16.0 ± 0.2 | 31.3 ± 0.4 | 13.7 ± 0.7 | 72.0 ± 5.1 | 344.8 ± 22.3 | 93.7 ± 0.5 | 18.0 ± 0.1 | 103.1 ± 5.3 |
| XLX/HZ F1 | 164.6 ± 1.5 | 18.0 ± 1.2 | 31.1 ± 0.4 | 14.4 ± 0.5 | 71.9 ± 4.5 | 334.3 ± 18.2 | 94.2 ± 0.1 | 21.0 ± 0.2 | 119.1 ± 6.4 |
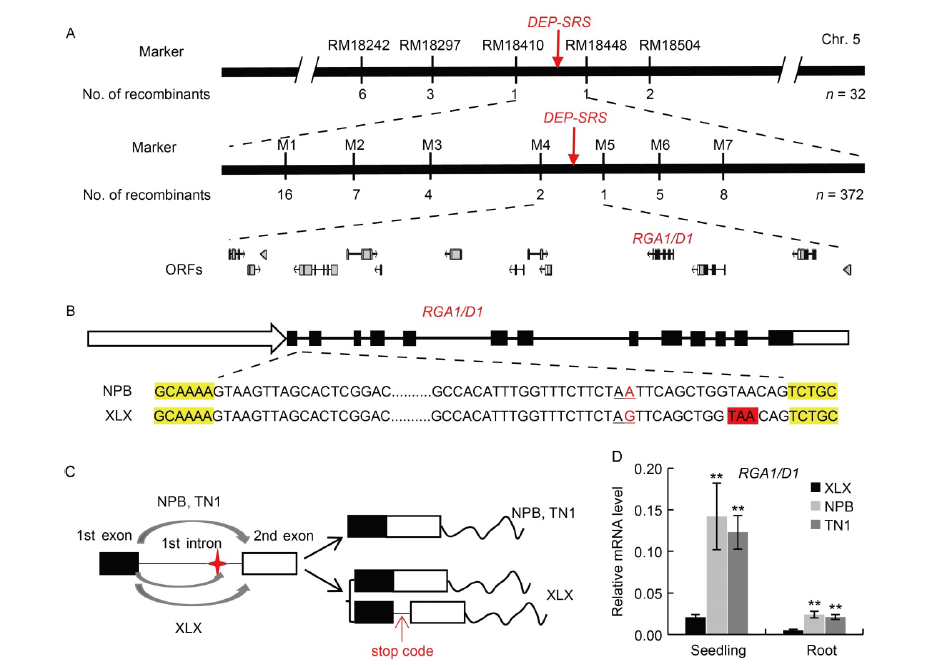
Fig. 2. Map-based cloning of DEP-SRS gene. A, Fine mapping of DEP-SRS gene. The gene was mapped to the interval between molecular markers RM18410 and RM18448 on chromosome (Chr.) 5 and further delimited to a 125-kb genomic region between markers M4 and M5 containing 14 open reading fragments (ORFs). Numbers below the markers indicate the number of recombinants. B, The first intron sequences of XLX and NPB. Black boxes indicate exons, black lines indicate introns, white box with arrow indicates promoter, and white box without arrow indicate 3′-UTR. Letters with yellow background indicate exon sequences, while those with red background indicate a stop code in the new transcript. Red letter with underline indicates the divergent sequence and black letter with underline indicates the splice site. C, The divergence (A to G) in the first intron produced a new splice site in XLX. Red star indicates the SNP, and red arrow indicates a stop code in the new transcript. D, Relative expression levels of functional RGA1/D1 transcripts. The two-week-old seedlings of XLX, NPB and TN1 were collected for qRT-PCR, and the OsACT gene was used as a control. Mean and SD values were obtained from three biological replicates. **, P < 0.01 (the Student’s t-test). XLX, Xiaolixiang; NPB, Nipponbare; TN1, Taichung Native 1; SNP, Single nucleotide polymorphism.
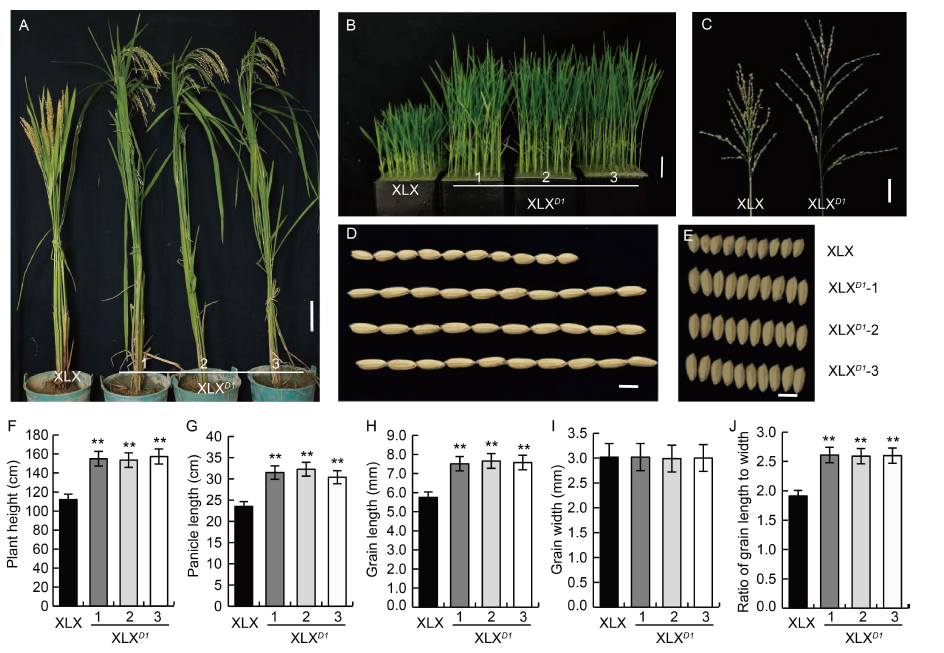
Fig. 3. Phenotypes of complementation transgenic lines of Xiaolixiang (XLXD1). A, Morphology at the maturity stage. Scale bar, 10 cm. B, Morphology at the seedling stage. Scale bar, 5 cm. C, Panicle type. Scale bar, 5 cm. D and E, Grain shape. Scale bars, 5 mm. F, Plant height. G, Panicle length. H, Grain length. I, Grain width. J, Ratio of grain length to width. Mean and SD values were obtained from three biological replicates. **, P < 0.01 (the Student’s t-test).
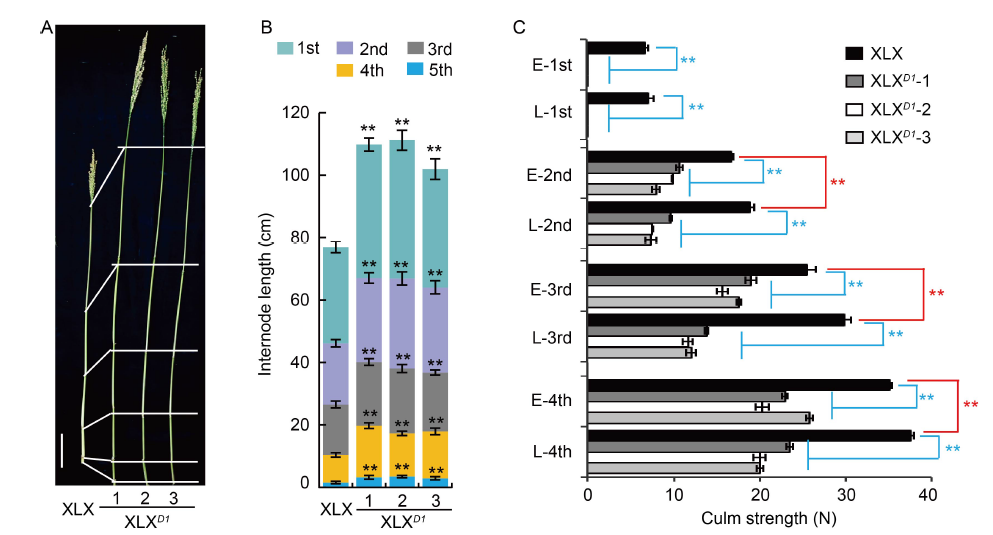
Fig. 4. Lodging resistance of Xiaolixiang (XLX) and its transgenic XLXD1 plants. A, Morphologies of culm and panicles. Scale bar, 10 cm. B, Internode length. C, Culm strengths of internodes at the early (E)- and late (L)-filling stages. 1st to 5th indicate the first internode to the last internode. Mean and SD values were obtained from three biological replicates. **, P < 0.01 (the Student’s t-test).
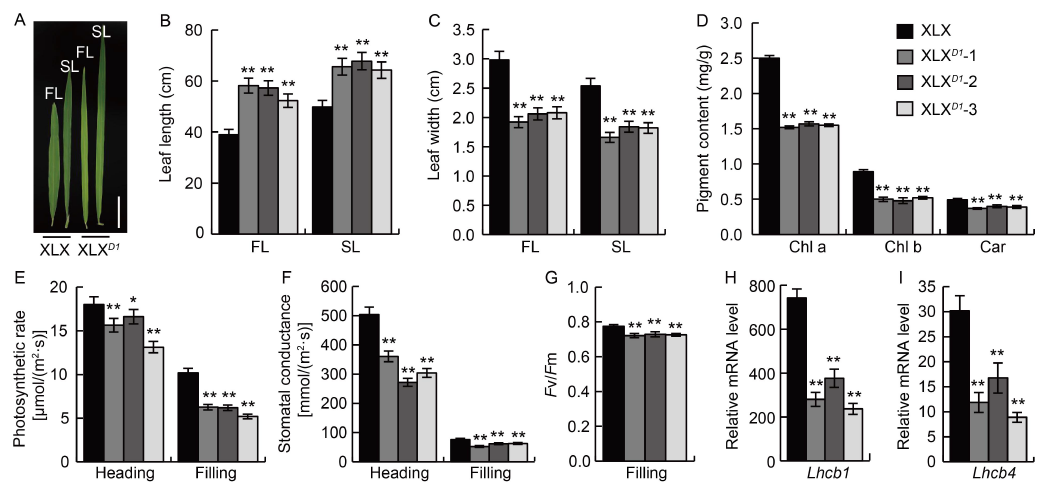
Fig. 5. Photosynthesis of Xiaolixiang (XLX) and its transgenic XLXD1 plants. A, Morphologies of flag leaf (FL) and secondary leaf (SL) at the filling stage. Scale bar, 10 cm. B, Leaf lengths of FL and SL at the filling stage. C, Leaf widths of FL and SL at the filling stage. D, Pigment content of FL at the filling stage. Chl a, Chlorophyll a; Chl b, Chlorophyll b; Car, Carotenoid. E and F, Photosynthetic rate (E) and stomatal conductance (F) of FL at the heading and filling stages. G, Optimal/maximal quantum yield of PS II (Fv/Fm) of FL at the filling stage. H and I, Relative expression levels of Lhcb1 and Lhcb4 in FL at the filling stage. Mean and SD values were obtained from three biological replicates.*, P < 0.05; **, P < 0.01 (the Student’s t-test).
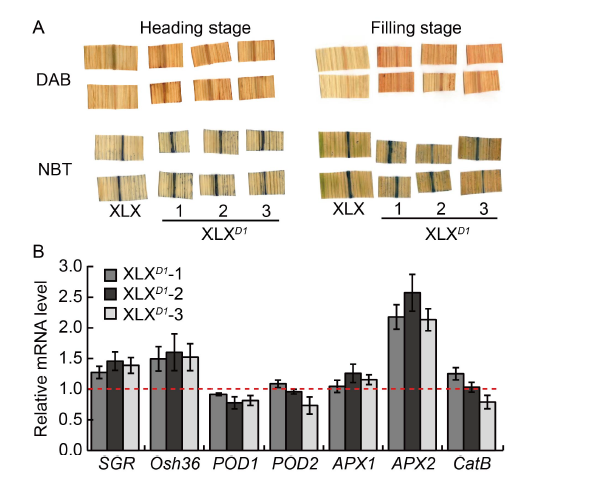
Fig. 6. Delayed leaf senescence in Xiaolixiang (XLX). A, Diaminobenzidine (DAB) staining and nitroblue tetrazolium (NBT) staining of leaves at the heading and filling stages. B, Changes in transcript levels of senescence-associated genes and ROS-scavenge enzyme genes in the leaves of XLX and XLXD1 plants at the filling stage. The flag leaves of XLX and XLXD1 plants at the filling stage were used for qRT-PCR. Two senescence-induced genes, SGR (LOC_Os09g36200) and Osh36 (LOC_Os05g39770); catalase gene CatB (LOC_Os06g51150); two peroxidase genes, POD1 (LOC_ Os01g22370) and POD2 (LOC_Os03g22010); two ascorbate peroxidase genes, APX1 (LOC_Os03g17690) and APX2 (LOC_Os07g49400) were detected, and the OsACT gene was used as a control. Mean and SD values were obtained from three biological replicates. The red line indicates the transcript level of these genes in XLX.
| Rice material | Effective panicle number per plant | Primary branch number per plant | Secondary branch number per plant | Grain number per spikelet | Seed-setting rate (%) | 1000-grain weight (g) | Theoretical yield per plant (g) |
|---|---|---|---|---|---|---|---|
| XLX | 15.8 ± 0.7 | 14.0 ± 0.5 | 53.6 ± 1.6 | 298.2 ± 19.5 | 90.6 ± 1.2 | 16.3 ± 0.1 | 70.1 ± 5.9 |
| XLXD1-1 | 11.0 ± 0.9** | 9.9 ± 0.6** | 28.4 ± 1.3** | 188.6 ± 6.1** | 85.9 ± 3.9 | 21.2 ± 0.2** | 37.7 ± 4.5** |
| XLXD1-2 | 11.0 ± 0.7** | 10.7 ± 0.2* | 34.7 ± 2.3* | 176.2 ± 8.6* | 87.4 ± 1.6 | 21.8 ± 0.1** | 36.9 ± 1.7** |
| XLXD1-3 | 10.7 ± 1.1** | 9.9 ± 0.4** | 25.9 ± 1.3* | 199.3 ± 13.0* | 86.1 ± 1.5 | 21.0 ± 0.2** | 38.7 ± 2.2** |
Table 2. Agronomic characteristics of Xiaolixiang (XLX) and its transgenic XLXD1 plants.
| Rice material | Effective panicle number per plant | Primary branch number per plant | Secondary branch number per plant | Grain number per spikelet | Seed-setting rate (%) | 1000-grain weight (g) | Theoretical yield per plant (g) |
|---|---|---|---|---|---|---|---|
| XLX | 15.8 ± 0.7 | 14.0 ± 0.5 | 53.6 ± 1.6 | 298.2 ± 19.5 | 90.6 ± 1.2 | 16.3 ± 0.1 | 70.1 ± 5.9 |
| XLXD1-1 | 11.0 ± 0.9** | 9.9 ± 0.6** | 28.4 ± 1.3** | 188.6 ± 6.1** | 85.9 ± 3.9 | 21.2 ± 0.2** | 37.7 ± 4.5** |
| XLXD1-2 | 11.0 ± 0.7** | 10.7 ± 0.2* | 34.7 ± 2.3* | 176.2 ± 8.6* | 87.4 ± 1.6 | 21.8 ± 0.1** | 36.9 ± 1.7** |
| XLXD1-3 | 10.7 ± 1.1** | 9.9 ± 0.4** | 25.9 ± 1.3* | 199.3 ± 13.0* | 86.1 ± 1.5 | 21.0 ± 0.2** | 38.7 ± 2.2** |
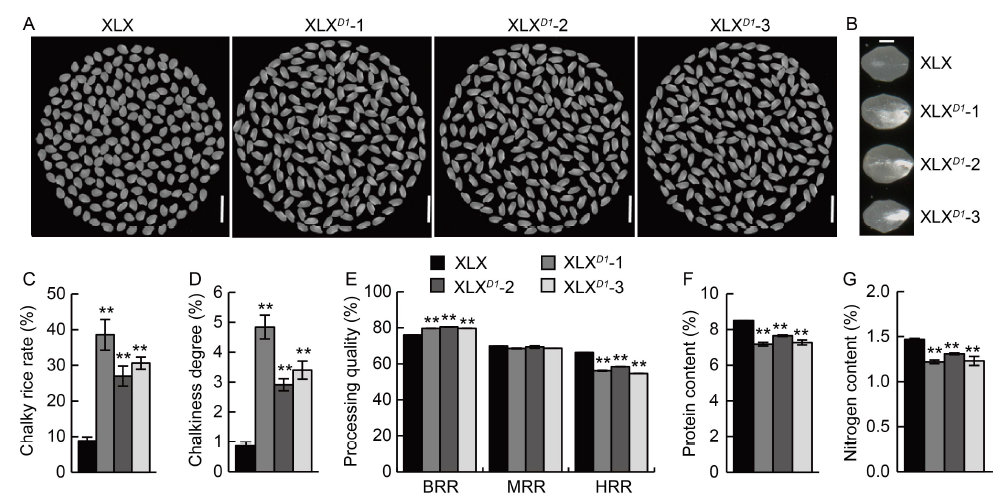
Fig. 7. Grain quality of Xiaolixiang (XLX) and its transgenic XLXD1 plants. A, Head rice of XLX and its transgenic XLXD1 lines (XLXD1-1, -2 and -3). Scale bars, 1 cm. B, Chalk sizes of XLX and XLXD1. Scale bar, 1 mm. C, Chalky rice rates of XLX and XLXD1. D, Chalkiness degrees of XLX and XLXD1. E, Processing qualities of XLX and XLXD1, including brown rice rate (BRR), milled rice rate (MRR) and head rice rate (HRR). F and G, Protein contents (F) and nitrogen contents (G) of XLX and XLXD1 head rice. Mean and SD values were obtained from three biological replicates. **, P < 0.01 (the Student’s t-test).
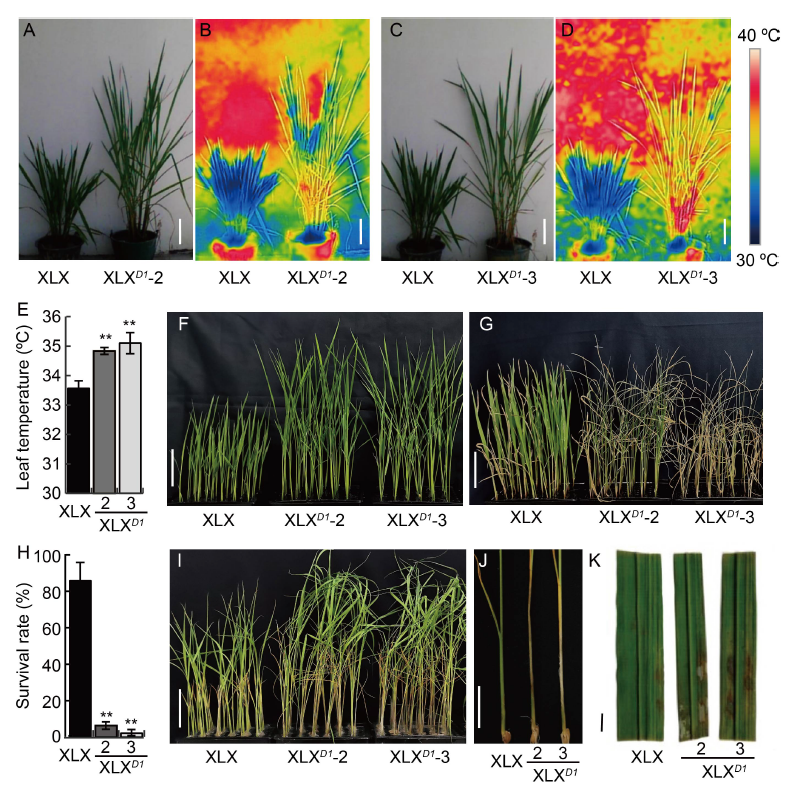
Fig. 8. Xiaolixiang (XLX) exhibits stronger drought and sheath blight resistances. A and C, Representative images of XLX, XLXD1-2 and XLXD1-3. Scale bars, 20 cm. B and D, Thermal images of XLX, XLXD1-2 and XLXD1-3. Scale bars, 20 cm. E, Leaf temperatures of XLX, XLXD1-2 and XLXD1-3 in paddy field. F-H, Seedlings of XLX, XLXD1-2 and XLXD1-3 before 20% polyethylene glycol (PEG) treatment (F), PEG treatment for 13 d (G) and survival rate after PEG treatment (H). Scale bars, 5 cm. I, Seedlings infected with Rhizoctonia solani. Scale bar, 5 cm. J, Lesion positions on seedlings after inoculation with R. solani. Scale bar, 2 cm. K, Adult leaves in vitro inoculated with R. solani. The leaves were collected at the tillering stage. Scale bar, 1 cm. Mean and SD values were obtained from three biological replicates. **, P < 0.01 (the Student’s t-test).
| [1] |
Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. 2012. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science, 335: 1348-1351.
PMID |
| [2] | Asano K, Miyao A, Hirochika H, Kitano H, Matsuoka M, Ashikari M. 2010. SSD1, which encodes a plant-specific novel protein, controls plant elongation by regulating cell division in rice. Proc Jpn Acad Ser B Phys Biol Sci, 86(3): 265-273. |
| [3] | Asano K, Yamasaki M, Takuno S, Miura K, Katagiri S, Ito T, Doi K, Wu J Z, Ebana K, Matsumoto T, Innan H, Kitano H, Ashikari M, Matsuoka M. 2011. Artificial selection for a green revolution gene during japonica rice domestication. Proc Natl Acad Sci USA, 108(27): 11034-11039. |
| [4] | Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. 1999. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc Natl Acad Sci USA, 96(18): 10284-10289. |
| [5] | Assmann S M. 2005. G protein regulation of disease resistance during infection of rice with rice blast fungus. Sci STKE, 2005: cm13. |
| [6] |
Caffarri S, Croce R, Cattivelli L, Bassi R. 2004. A look within LHCII: Differential analysis of the Lhcb1-3 complexes building the major trimeric antenna complex of higher-plant photosynthesis. Biochemistry, 43(29): 9467-9476.
PMID |
| [7] | Chen W W, Cheng Z J, Liu L L, Wang M, You X M, Wang J, Zhang F, Zhou C L, Zhang Z, Zhang H, You S M, Wang Y P, Luo S, Zhang J H, Wang J L, Wang J, Zhao Z C, Guo X P, Lei C L, Zhang X, Lin Q B, Ren Y L, Zhu S S, Wan J M. 2019. Small Grain and Dwarf 2, encoding an HD-Zip II family transcription factor, regulates plant development by modulating gibberellin biosynthesis in rice. Plant Sci, 288: 110208. |
| [8] |
Cui Y, Jiang N, Xu Z J, Xu Q. 2020. Heterotrimeric G protein are involved in the regulation of multiple agronomic traits and stress tolerance in rice. BMC Plant Biol, 20(1): 90.
PMID |
| [9] | Feng B H, Yang Y, Shi Y F, Lin L, Chen J, Wei Y L, Leung H, Wu J L. 2013. Genetic analysis and gene mapping of light brown spotted leaf mutant in rice. Rice Sci, 20(1): 13-18. |
| [10] |
Ferrero-Serrano Á, Assmann S M. 2016. The α-subunit of the rice heterotrimeric G protein, RGA1, regulates drought tolerance during the vegetative phase in the dwarf rice mutant d1. J Exp Bot, 67(11): 3433-3443.
PMID |
| [11] | Ferrero-Serrano Á, Su Z, Assmann S M. 2018. Illuminating the role of the Gα heterotrimeric G protein subunit, RGA1, in regulating photoprotection and photoavoidance in rice. Plant Cell Environ, 41(2): 451-468. |
| [12] | Ferrero-Serrano Á, Cantos C, Assmann S M. 2019. The role of dwarfing traits in historical and modern agriculture with a focus on rice. Cold Spring Harb Perspect Biol, 11(11): a034645. |
| [13] | Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y. 1999. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA, 96(13): 7575-7580. |
| [14] |
Gilman A G. 1987. G proteins: Transducers of receptor-generated signals. Annu Rev Biochem, 56: 615-649.
PMID |
| [15] |
Hedden P. 2003. The genes of the green revolution. Trends Genet, 19(1): 5-9.
PMID |
| [16] | Hong Y B, Zhang Y X, Sinumporn S, Yu N, Zhan X D, Shen X H, Chen D B, Yu P, Wu W X, Liu Q E, Cao Z L, Zhao C D, Cheng S H, Cao L Y. 2018. Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice. Plant J, 95: 877-891. |
| [17] |
Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, Uozu S, Kitano H, Ashikari M, Matsuoka M. 2002. Loss-of- function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J, 32(4): 495-508.
PMID |
| [18] | Hu J, Zhu L, Zeng D L, Gao Z Y, Guo L B, Fang Y X, Zhang G H, Dong G J, Yan M X, Liu J, Qian Q. 2010. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol Biol, 73(3): 283-292. |
| [19] |
Khush G S. 2001. Green revolution: The way forward. Nat Rev Genet, 2(10): 815-822.
PMID |
| [20] | Li M R, Li H Q. 2003. A simple and highly efficient Agrobacterium- mediated rice transformation system. Acta Biol Exp Sin, 36(4): 289-294. (in Chinese with English abstract) |
| [21] | Li S, Tian Y H, Wu K, Ye Y F, Yu J P, Zhang J Q, Liu Q, Hu M Y, Li H, Tong Y P, Harberd N P, Fu X D. 2018. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature, 560: 595-600. |
| [22] | Liu C, Li L G. 2016. Advances in molecular understanding of rice lodging resistance. Chin J Rice Sci, 30(2): 216-222. (in Chinese with English abstract) |
| [23] |
Liu F, Wang P D, Zhang X B, Li X F, Yan X H, Fu D H, Wu G. 2018. The genetic and molecular basis of crop height based on a rice model. Planta, 247(1): 1-26.
PMID |
| [24] |
Liu X, Hu Q L, Yan J J, Sun K, Liang Y, Jia M R, Meng X B, Fang S, Wang Y Q, Jing Y H, Liu G F, Wu D X, Chu C C, Smith S M, Chu J F, Wang Y H, Li J Y, Wang B. 2020. ζ-carotene isomerase suppresses tillering in rice through the coordinated biosynthesis of strigolactone and abscisic acid. Mol Plant, 13(12): 1784-1801.
PMID |
| [25] | Miura K, Agetsuma M, Kitano H, Yoshimura A, Matsuoka M, Jacobsen S E, Ashikari M. 2009. A metastable DWARF1 epigenetic mutant affecting plant stature in rice. Proc Natl Acad Sci USA, 106(27): 11218-11223. |
| [26] |
Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M S, Sato M, Nasu S, Minobe Y. 2002. Positional cloning of rice semidwarfing gene, sd-1: Rice ‘green revolution gene’ encodes a mutant enzyme involved in gibberellin synthesis. DNA Res, 9(1): 11-17.
PMID |
| [27] | Murai M, Hirose S, Sato S, Takebe M. 1991. Effects of dwarfing genes from dee-geo-woo-gen and other varieties on cool temperature tolerance at booting stage in rice. Jpn J Breed, 41(2): 241-254. |
| [28] |
Murchie E H, Yang J C, Hubbart S, Horton P, Peng S B. 2002. Are there associations between grain-filling rate and photosynthesis in the flag leaves of field-grown rice? J Exp Bot, 53: 2217-2224.
PMID |
| [29] | Oki K, Inaba N, Kitagawa K, Fujioka S, Kitano H, Fujisawa Y, Kato H, Iwasaki Y. 2009. Function of the α subunit of rice heterotrimeric G protein in brassinosteroid signaling. Plant Cell Physiol, 50: 161-172. |
| [30] |
Pathak R R, Mandal V K, Jangam A P, Sharma N, Madan B, Jaiswal D K, Raghuram N. 2021. Heterotrimeric G-protein α subunit (RGA1) regulates tiller development, yield, cell wall, nitrogen response and biotic stress in rice. Sci Rep, 11: 2323.
PMID |
| [31] | Peng P, Gao Y D, Li Z, Yu Y W, Qin H, Guo Y, Huang R F, Wang J. 2019. Proteomic analysis of a rice mutant sd58 possessing a novel d1 allele of heterotrimeric G protein alpha subunit (RGA1) in salt stress with a focus on ROS scavenging. Int J Mol Sci, 20(1): 167. |
| [32] | Porra R J, Schäfer W, Cmiel E, Katheder I, Scheer H. 1994. The derivation of the formyl-group oxygen of chlorophyll b in higher plants from molecular oxygen. Eur J Biochem, 219(1/2): 671-679. |
| [33] | Qi W W, Sun F, Wang Q J, Chen M L, Huang Y Q, Feng Y Q, Luo X J, Yang J S. 2011. Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol, 157(1): 216-228. |
| [34] | Sazuka T, Kamiya N, Nishimura T, Ohmae K, Sato Y, Imamura K, Nagato Y, Koshiba T, Nagamura Y, Ashikari M, Kitano H, Matsuoka M. 2009. A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J, 60(2): 227-241. |
| [35] | Sha H J, Liu H L, Zhao G X, Han Z M, Chang H L, Wang J G, Zheng H L, Zhang J F, Yu Y, Liu Y Q, Zou D T, Nie S J, Fang J. 2022. Elite sd1 alleles in japonica rice and their breeding applications in northeast China. Crop J, 10(1): 224-233. |
| [36] | Suharsono U, Fujisawa Y, Kawasaki T, Iwasaki Y, Satoh H, Shimamoto K. 2002. The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA, 99(20): 13307-13312. |
| [37] |
Sun H Y, Qian Q, Wu K, Luo J J, Wang S S, Zhang C W, Ma Y F, Liu Q, Huang X Z, Yuan Q B, Han R X, Zhao M, Dong G J, Guo L B, Zhu X D, Gou Z H, Wang W, Wu Y J, Lin H X, Fu X D. 2014. Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat Genet, 46(6): 652-656.
PMID |
| [38] | Sun Q, Yang S, Guo X F, Wang S T, Jia X T, Li S, Xuan Y H. 2021. RAVL1 activates IDD3 to negatively regulate rice resistance to sheath blight disease. Rice Sci, 28(2): 146-155. |
| [39] | Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M. 2000. Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA, 97(21): 11638-11643. |
| [40] |
Urano D, Colaneri A, Jones A M. 2014. Gα modulates salt-induced cellular senescence and cell division in rice and maize. J Exp Bot, 65(22): 6553-6561.
PMID |
| [41] |
Vikram P, Swamy B P M, Dixit S, Singh R, Singh B P, Miro B, Kohli A, Henry A, Singh N K, Kumar A. 2015. Drought susceptibility of modern rice varieties: An effect of linkage of drought tolerance with undesirable traits. Sci Rep, 5: 14799.
PMID |
| [42] | Wang F M, Yoshida H, Matsuoka M. 2021. Making the ‘green revolution’ truly green: Improving crop nitrogen use efficiency. Plant Cell Physiol, 62(6): 942-947. |
| [43] |
Wang L, Xu Y Y, Ma Q B, Li D, Xu Z H, Chong K. 2006. Heterotrimeric G protein alpha subunit is involved in rice brassinosteroid response. Cell Res, 16(12): 916-922.
PMID |
| [44] | Wang R N, Liu C, Li Q Z, Chen Z N, Sun S Y, Wang X L. 2020. Spatiotemporal resolved leaf angle establishment improves rice grain yield via controlling population density. iScience, 23(9): 101489. |
| [45] | Wang Z B, Zuo S M, Li G, Chen X J, Chen Z X, Zhang Y F, Pan X B. 2009. Rapid identification technology of resistance to rice sheath blight in seedling stage. Acta Phytopath Sin, 39: 174-182. (in Chinese with English abstract) |
| [46] | Yaish M W, El-Kereamy A, Zhu T, Beatty P H, Good A G, Bi Y M, Rothstein S J. 2010. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet, 6(9): e1001098. |
| [47] |
Yang Y L, Xu J, Huang L C, Leng Y J, Dai L P, Rao Y C, Chen L, Wang Y Q, Tu Z J, Hu J, Ren D Y, Zhang G H, Zhu L, Guo L B, Qian Q, Zeng D L. 2016. PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J Exp Bot, 67(5): 1297-1310.
PMID |
| [48] | Yuan L P. 2014. Development of hybrid rice to ensure food security. Rice Sci, 21(1): 1-2. |
| [49] | Zhu Y C, Li T, Xu J, Wang J J, Wang L, Zou W W, Zeng D L, Zhu L, Chen G, Hu J, Gao Z Y, Dong G J, Ren D Y, Shen L, Zhang Q, Guo L B, Hu S P, Qian Q, Zhang G H. 2020. Leaf width gene LW5/D1 affects plant architecture and yield in rice by regulating nitrogen utilization efficiency. Plant Physiol Biochem, 157: 359-369. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [13] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| [14] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| [15] | Zhang Guomei, Li Han, Liu Shanshan, Zhou Xuming, Lu Mingyang, Tang Liang, Sun Lihua. Water Extract of Rice False Smut Balls Activates Nrf2/HO-1 and Apoptosis Pathways, Causing Liver Injury [J]. Rice Science, 2023, 30(5): 473-485. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||