
Rice Science ›› 2019, Vol. 26 ›› Issue (6): 372-383.DOI: 10.1016/j.rsci.2019.03.001
• Research Paper • Previous Articles Next Articles
Ting Chen, Zheng Chen, Prakash Sathe Atul, Zhihong Zhang, Liangjian Li, Huihui Shang, Shaoqing Tang, Xiaobo Zhang( ), Jianli Wu(
), Jianli Wu( )
)
Received:2019-03-27
Accepted:2019-05-31
Online:2019-11-28
Published:2019-08-19
Ting Chen, Zheng Chen, Prakash Sathe Atul, Zhihong Zhang, Liangjian Li, Huihui Shang, Shaoqing Tang, Xiaobo Zhang, Jianli Wu. Characterization of a Novel Gain-of-Function Spotted-Leaf Mutant with Enhanced Disease Resistance in Rice[J]. Rice Science, 2019, 26(6): 372-383.
Add to citation manager EndNote|Ris|BibTeX
| Marker | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|
| ACTIN | AGGCTCCTCTCAACCCCAAG | TTTCCTGGTCATAGTCCAGG |
| OsPR10 | CACCATCTACACCATGAAGC | AGCACATCCGACTTTAGGAC |
| OsLOX | GATGGCGGTGCTCGACGTGCT | GCACCTGTTCTTGAGCTTTCTAT |
| OsAOS2 | CTCGTCGGAAGGCTGTTGCT | ACGATTGACGGCGGAGGTT |
| OsJamyb | CCGAGCATGGTGACTAGCTCATCTT | CCTTGCACCCAACCGTTAAGCTGTT |
| OsWRKY45 | TTCCTTGTTGATGTGTCGTCTCA | CCCCCAGCTCATAATCAAGAAC |
| OsNPR1 | GGCAGGTGAGAGTCTACGAGGAA | GCTGTCATCCGAGCTAAGTGTT |
| OsEDS1 | CATTCCAAGAACGAGGACACTG | CAAGACTCAAGGCTAGAACCGA |
| OsPAL3 | CGCTGAGGCGTTTAAGATTG | GGCAAGGACAGCAAGAATG |
| OsPAL4 | CTTCACAACAGCTAATCGAG | CGCACTCCATTTCAGTACCA |
Supplemental Table 1 List of primers for qRT-PCR.
| Marker | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|
| ACTIN | AGGCTCCTCTCAACCCCAAG | TTTCCTGGTCATAGTCCAGG |
| OsPR10 | CACCATCTACACCATGAAGC | AGCACATCCGACTTTAGGAC |
| OsLOX | GATGGCGGTGCTCGACGTGCT | GCACCTGTTCTTGAGCTTTCTAT |
| OsAOS2 | CTCGTCGGAAGGCTGTTGCT | ACGATTGACGGCGGAGGTT |
| OsJamyb | CCGAGCATGGTGACTAGCTCATCTT | CCTTGCACCCAACCGTTAAGCTGTT |
| OsWRKY45 | TTCCTTGTTGATGTGTCGTCTCA | CCCCCAGCTCATAATCAAGAAC |
| OsNPR1 | GGCAGGTGAGAGTCTACGAGGAA | GCTGTCATCCGAGCTAAGTGTT |
| OsEDS1 | CATTCCAAGAACGAGGACACTG | CAAGACTCAAGGCTAGAACCGA |
| OsPAL3 | CGCTGAGGCGTTTAAGATTG | GGCAAGGACAGCAAGAATG |
| OsPAL4 | CTTCACAACAGCTAATCGAG | CGCACTCCATTTCAGTACCA |
| Marker | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|
| RM6697 | TATTCCCGGGAGATCCAACAGC | AAGATCCAGTCGATTTGGTTCAGG |
| RM5752 | TTGCAATTAATTCGATCTCC | GCAGATCGATTCGTTAGTTC |
| RM5490 | GCGGTGTGATTTGAATTGAACTGG | GCAAAGCAACTTCAAAGCTTCTCG |
| InDel42 | ACAAATCTAGAACAGGTGGCA | GGAATGCTCACGTACGTTCAA |
Supplemental Table 2 List of primers for gene mapping.
| Marker | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|
| RM6697 | TATTCCCGGGAGATCCAACAGC | AAGATCCAGTCGATTTGGTTCAGG |
| RM5752 | TTGCAATTAATTCGATCTCC | GCAGATCGATTCGTTAGTTC |
| RM5490 | GCGGTGTGATTTGAATTGAACTGG | GCAAAGCAACTTCAAAGCTTCTCG |
| InDel42 | ACAAATCTAGAACAGGTGGCA | GGAATGCTCACGTACGTTCAA |
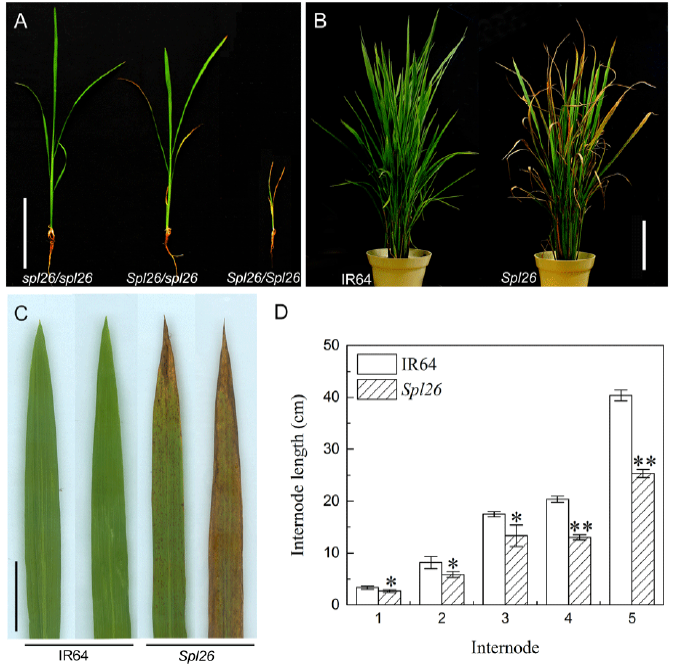
Fig. 1. Phenotypes of Spl26 and the wild-type IR64.A, Phenotypes of Spl26 plants grown under normal field conditions for 19 d. spl26/spl26 is a wild-type, Spl26/spl26 is a heterozygous plant, and Spl26/Spl26 is a homozygous plant. Scale bar = 20 cm. B, IR64 and Spl26 at the tillering stage. Scale bar = 20 cm. C, Leaves of Spl26 and IR64. Scale bar = 2 cm. D, Internode length of IR64 and Spl26.Values are Mean ± SD (n = 3). * and ** indicate significant differences at the 0.05 and 0.01 levels by the Student’s t test, respectively.
| Material | Plant height (cm) | Panicle length (cm) | No. of tillers per plant | No. of filled grains per panicle | Seed-setting rate (%) | 1000-grain weight (g) |
|---|---|---|---|---|---|---|
| IR64 | 116.67 ± 1.53 | 27.50 ± 0.50 | 19.67 ± 1.53 | 80.67 ± 3.79 | 60.04 ± 1.71 | 24.63 ± 0.40 |
| Spl26 | 78.00 ± 1.73** | 18.00 ± 0.50** | 14.00 ± 1.00** | 26.67 ± 1.53** | 47.34 ± 2.37** | 19.47 ± 0.21** |
Table 1 Comparison of agronomic traits between IR64 and Spl26.
| Material | Plant height (cm) | Panicle length (cm) | No. of tillers per plant | No. of filled grains per panicle | Seed-setting rate (%) | 1000-grain weight (g) |
|---|---|---|---|---|---|---|
| IR64 | 116.67 ± 1.53 | 27.50 ± 0.50 | 19.67 ± 1.53 | 80.67 ± 3.79 | 60.04 ± 1.71 | 24.63 ± 0.40 |
| Spl26 | 78.00 ± 1.73** | 18.00 ± 0.50** | 14.00 ± 1.00** | 26.67 ± 1.53** | 47.34 ± 2.37** | 19.47 ± 0.21** |

Fig. 2. Effect of light on the formation of lesion in Spl26.A, IR64 before shading. B, Spl26 before shading. C, IR64 shaded for 3 d. D, Spl26 shaded for 3 d. E, Spl26 reinstated for 7 d. F, Spl26 leaf with lesions shaded for 7 d. Shaded areas are boxed. Scale bar = 2 cm.

Fig. 3. Photosynthetic pigment contents and photosynthetic parameters of Spl26 and IR64.A, Photosynthetic pigment contents of Spl26 and IR64 at the tillering stage. B, Photosynthetic pigment contents of Spl26 and IR64 at the heading stage. C, Net photosynthetic rate (Pn). D, Stomatal conductance (Gs). E, Transpiration rate (Tr). F, Intercellular CO2 concentration (Ci).Values are Mean ± SD (n = 3). * and ** indicate significant differences at the 0.05 and 0.01 levels by the Student’s t test, respectively.
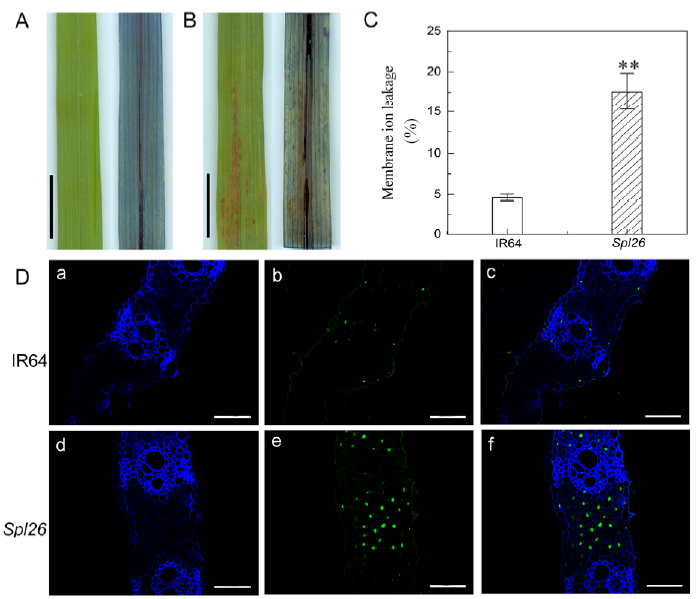
Fig. 4. Determination of cell death indicators in Spl26 and IR64.A, Trypan blue staining of IR64 at the tillering stage. Scale bar = 1 cm. B, Trypan blue staining of Spl26 at the tillering stage. Scale bar = 1 cm. C, Membrane ion leakage rate at the heading stage. D, Terminal deoxyribonucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay at the tillering stage. Blue signal represents 40,6-diamino- phenylindole (DAPI). Green color represents positive result. (a) and (d) are DAPI staining; (b) and (e) are TUNEL signal; (c) and (f) are merged images of (a/b) and (d/e), respectively. Scale bar = 50 μm.Values are Mean ± SD (n = 3). ** indicates significant differences at P ≤ 0.01 by the Student’s t test.
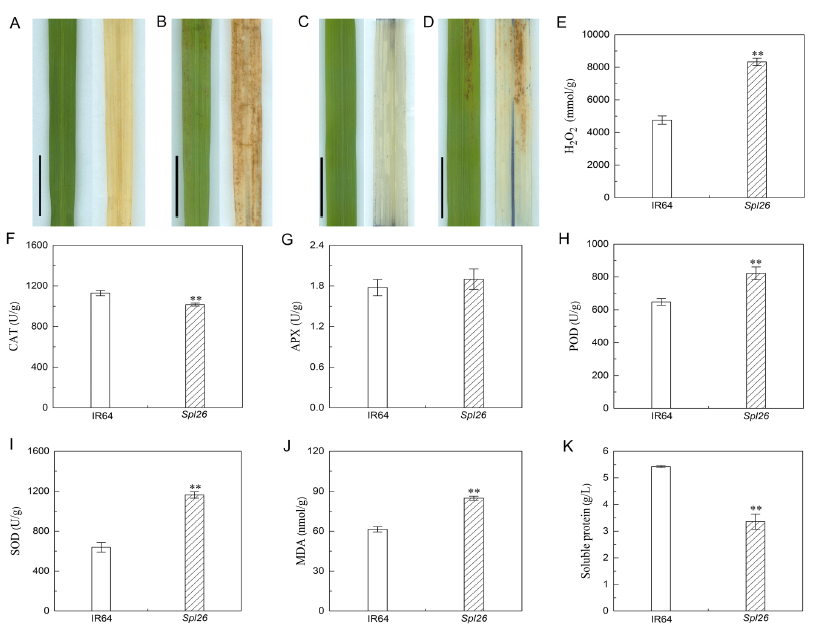
Fig. 5. Determination of reactive oxygen species (ROS)-associated parameters in Spl26 and IR64.A, 3,3'-diaminobenzidine (DAB) staining of IR64 at the tillering stage. Scale bar = 2 cm. B, DAB staining of Spl26 at the tillering stage. Scale bar = 2 cm. C, Nitroblue tetrazolium (NBT) staining of IR64 at the tillering stage. Scale bar = 2 cm. D, NBT staining of Spl26 at the tillering stage. Scale bar = 2 cm. E, H2O2 content. F, Catalase (CAT) activity. G, Ascorbate peroxidase (APX) activity. H, Peroxidase (POD) activity. I, Superoxide dismutase (SOD) activity. J, Malonaldehyde (MDA) content. K, Soluble protein content.Values are Mean ± SD (n = 3). ** indicates significant difference at the 0.01 level by the Student’s t test.
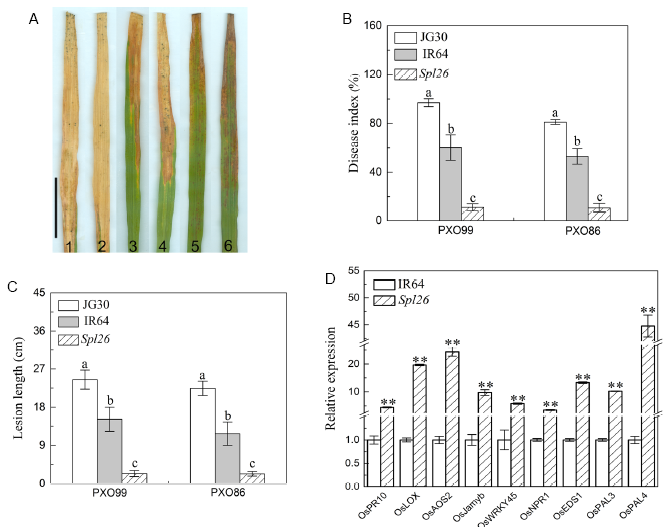
Fig. 6. Evaluation of disease resistance and expression of defense response genes in Spl26 and IR64.A, Reaction to PXO99. 1-2, JG30; 3-4, IR64; 5-6, Spl26. Scale bar = 5 cm. B, Disease index (lesion length/leaf length ratio). Different lowercase letters above the bars indicate difference at P ≤ 0.05 by the Duncan’s test. C, Lesion length. Different letters indicate a statistical difference at P ≤ 0.05 by the Duncan’s test. D, Expression of defense response genes in Spl26 and IR64. The expression level of each gene in IR64 was normalized to 1. ** indicates significant difference at P ≤ 0.01 by the Student’s t test.Values are Mean ± SD (n = 3).
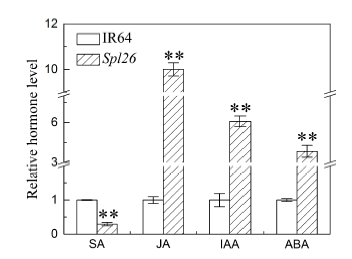
Fig. 7. Contents of hormones in Spl26 and IR64.SA, Salicylic acid; JA, Jasmonic acid; IAA, Indole acetic acid; ABA, Abscisic acid.Hormone level in IR64 was normalized to 1. Values are Mean ± SD (n = 3). ** indicates significant difference at P ≤ 0.01 by the Student’s t test.
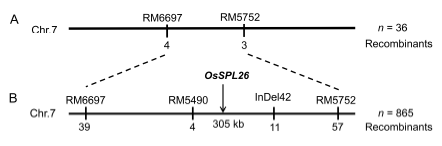
Fig. 8. Map-based isolation of OsSPL26.A, OsSPL26 was located on the short arm of chromosome 7 between RM6697 and RM5752. B, Fine mapping of OsSPL26 based on 865 wild-type F2 individuals.
| [1] | Arnon D I.1949. Copper enzymes in isolated chloroplasts: Polyphenoloxidase inBeta vulgaris. Plant Physiol, 24(1): 1-15. |
| [2] | Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Töpsch S, Vos P, Salamini F, Schulze-Lefert P.1997. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell, 88(5): 695-705. |
| [3] | Chen Z, Chen T, Sathe A P, He Y Q, Zhang X B, Wu J L.2018. Identification of a novel semi-dominant spotted-leaf mutant with enhanced resistance to Xanthomonas oryzae pv. oryzae in rice. Int J Mol Sci, 19(12): 3766. |
| [4] | Dietrich R A, Delaney T P, Uknes S J, Ward E R, Ryals J A, Dangl J L.1994. Arabidopsis mutants simulating disease resistance response. Cell, 77(4): 565-577. |
| [5] | Dietrich R A, Richberg M H, Schmidt R, Dean C, Dangl J L.1997. A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell, 88(5): 685-694. |
| [6] | Edwards K, Johnstone C, Thompson C.1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis.Nucl Acids Res, 19(6): 1349. |
| [7] | Feng B H, Yang Y, Shi Y F, Shen H C, Wang H M, Huang Q N, Xu X, Lü X G, Wu J L.2013. Characterization and genetic analysis of a novel rice spotted-leaf mutant HM47 with broad-spectrum resistance to Xanthomonas oryzae pv. oryzae. J Integr Plant Biol, 55(5): 473-483. |
| [8] | Fu Z Q, Yan S P, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel S H, Tada Y, Zheng N, Dong X N.2012. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants.Nature, 486: 228-232. |
| [9] | Gray J, Close P S, Briggs S P, Johal G S.1997. A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell, 89(1): 25-31. |
| [10] | Greenberg J T, Guo A, Klessig D F, Ausubel F M.1994. Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions.Cell, 77(4): 551-563. |
| [11] | Greenberg J T.1997. Programmed cell death in plant-pathogen interactions.Annu Rev Plant Biol, 48: 525-545. |
| [12] | He Y, Zhang Z H, Li L J, Tang S Q, Wu J L.2018. Genetic and physio-biochemical characterization of a novel premature senescence leaf mutant in rice (Oryza sativa L.). Int J Mol Sci, 19(8): 2339. |
| [13] | Hu G S, Yalpani N, Briggs S P, Johal G S.1998. A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize.Plant Cell, 10(7): 1095-1105. |
| [14] | Huang Q N, Yang Y, Shi Y F, Chen J, Wu J L.2010. Spotted-leaf mutants of rice (Oryza sativa). Rice Sci, 17(4): 247-256. |
| [15] | Huang Q N, Shi Y F, Yang Y, Feng B H, Wei Y L, Chen J, Baraoidan M, Leung H, Wu J L.2011. Characterization and genetic analysis of a light- and temperature-sensitive spotted-leaf mutant in rice.J Integr Plant Biol, 53(8): 671-681. |
| [16] | Huang Q N, Shi Y F, Zhang X B, Song L X, Feng B H, Wang H M, Xu X, Li X H, Guo D, Wu J L.2016. Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J Integr Plant Biol, 58(1): 12-28. |
| [17] | Jiao B B, Wang J J, Zhu X D, Zeng L J, Li Q, He Z H.2012. A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice.Mol Plant, 5(1): 205-217. |
| [18] | Kauffman H E, Reddy A P K, Hsieh S P V, Merca S D.1973. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep, 57: 537-541. |
| [19] | Kiyosawa S.1970. Inheritance of a particular sensitivity of the rice variety, Sekiguchi-Asahi, to pathogens and chemicals, and linkage relationship with blast resistance.Bull Nat Inst Agric Sci (Jpn) Ser D: Physiol Genet, 21: 61-71. |
| [20] | Li L F, Xiong Y Y, Ouyang L J, Peng X S, Chen X R, He X P, Fu J R, Bian J M, Hu L F, Xu J, He H H, Sun X T, Zhu C L.2018. Identification and gene mapping of white stripe leaf and white panicle mutantwlp6 in rice. Chin J Rice Sci, 32(6): 538-548. (in Chinese with English abstract) |
| [21] | Li X Z, Yang D L, Sun L, Li Q, Mao B Z, He Z H.2016. The systemic acquired resistance regulatorOsNPR1 attenuates growth by repressing auxin signaling through promoting IAA- amido synthase expression. Plant Physiol, 172(1): 546-558. |
| [22] | Lim P O, Kim H J, Nam H G.2007. Leaf senescence.Annu Rev Plant Biol, 58: 115-136. |
| [23] | Liu Q E, Ning Y S, Zhang Y X, Yu N, Zhao C D, Zhan X D, Wu W X, Chen D B, Wei X J, Wang G L, Cheng S H, Cao L Y.2017. OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell, 29(2): 345-359. |
| [24] | Liu X Q, Li F, Tang J Y, Wang W H, Zhang F X, Wang G D, Chu J F, Yan C Y, Wang T Q, Chu C C, Li C Y.2012. Activation of the jasmonic acid pathway by depletion of the hydroperoxide lyase OsHPL3 reveals crosstalk between the HPL and AOS branches of the oxylipin pathway in rice.PLoS One, 7(11): e50089. |
| [25] | Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R.2010. Reactive oxygen species homeostasis and signaling during drought and salinity stresses.Plant Cell Environ, 33(4): 453-467. |
| [26] | Mizobuchi R, Hirabayashi H, Kaji R, Nishizawa Y, Satoh H, Ogawa T, Okamoto M.2002. Differential expression of disease resistance in rice lesion-mimic mutants.Plant Cell Rep, 21(4): 390-396. |
| [27] | Mori M, Tomita C, Sugimoto K, Hasegawa M, Hayashi N, Dubouzet J G, Ochiai H, Sekimoto H, Hirochika H, Kikuchi S.2007. Isolation and molecular characterization of aspotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol Biol, 63(6): 847-860. |
| [28] | Qiao Y L, Jiang W Z, Lee J H, Park B S, Choi M S, Piao R H, Woo M O, Roh J H, Han L Z, Paek N C, Seo H S, Koh H J.2010. SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit μ1 (AP1M1) and is responsible for spotted leaf and early senescence in rice(Oryza sativa). New Phytol, 185(1): 258-274. |
| [29] | Salt J N, Yoshioka K, Moeder W, Goring D R.2011. Altered germination and subcellular localization patterns for PUB44/ SAUL1 in response to stress and phytohormone treatments.PLoS One, 6(6): e21321. |
| [30] | Schlimme M, Blaschke L, Lagrimini L M, Polle A.2002. Growth performance and lignification in tobacco with suppressed apoplastic anionic peroxidase activity under ambient and elevated CO2 concentrations.Intl J Plant Sci, 163(5): 749-754. |
| [31] | Shen X L, Liu H B, Yuan B, Li X H, Xu C G, Wang S P.2011. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ, 34(2): 179-191. |
| [32] | Song G, Kwon C T, Kim S H, Shim Y, Lim C, Koh H J, An G, Kang K, Paek N C.2019. The rice SPOTTED LEAF4 (SPL4) encodes a plant spastin that inhibits ROS accumulation in leaf development and functions in leaf senescence. Front Plant Sci, 9: 1925. |
| [33] | Sparkes I A, Brandizzi F, Slocombe S P, El-Shami M, Hawes C, Baker A.2003. An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol, 133(4): 1809-1819. |
| [34] | Strecker V, Mai S, Muster B, Beneke S, Bürkle A, Bereiter-Hahn J, Jendrach M.2010. Aging of different avian cultured cells: Lack of ROS-induced damage and quality control mechanisms.Mech Ageing Dev, 131(1): 48-59. |
| [35] | Sun L T, Wang Y H, Liu L L, Wang C M, Gan T, Zhang Z Y, Wang Y L, Wang D, Niu M, Long W H, Li X H, Zheng M, Jiang L, Wan J M.2017a. Isolation and characterization of a leaf spotted leaf 32 mutant with early leaf senescence and enhanced defense response in rice.Sci Rep, 7: 41846. |
| [36] | Sun L T, Lin T Z, Wang Y L, Niu M, Hu T T, Liu S J, Wang Y H, Wan J M.2017b. Phenotypic analysis and gene mapping of a white stripe mutant st13 in rice. Chin J Rice Sci, 31(4): 335-363. (in Chinese with English abstract) |
| [37] | Takahashi A, Kawasaki T, Henmi K, Shii K, Kodama O, Satoh H, Shimamoto K.1999. Lesion mimic mutants of rice with alterations in early signaling events of defense.Plant J, 17(5): 535-545. |
| [38] | Tang J Y, Zhu X D, Wang Y Q, Liu L C, Xu B, Li F, Fang J, Chu C C.2011. Semi-dominant mutations in the CC-NB-LRR-typeR gene, NLS1, lead to constitutive activation of defense responses in rice. Plant J, 66(6): 996-1007. |
| [39] | Thordal-Christensen H, Zhang Z G, Wei Y D, Collinge D B.1997. Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction.Plant J, 11(6): 1187-1194. |
| [40] | Vijayan P, Shockey J, Levesque C A, Cook R J, Browse J.1998. A role for jasmonate in pathogen defense of Arabidopsis. Proc Natl Acad Sci USA, 95: 7209-7214. |
| [41] | Wang S, Lei C L, Wang J L, Ma J, Tang S, Wang C L, Zhao K J, Tian P, Zhang H, Qi C Y, Cheng Z J, Zhang X, Guo X P, Liu L L, Wu C Y, Wan J M.2017. SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J Exp Bot, 68(5): 899-913. |
| [42] | Wang S H, Lim J H, Kim S S, Cho S H, Yoo S C, Koh H J, Sakuraba Y, Paek N C.2015. Mutation of SPOTTED LEAF3 (SPL3) impairs abscisic acid responsive signaling and delays leaf senescence in rice. J Exp Bot, 66(22): 7045-7059. |
| [43] | Wang Z H, Wang Y, Hong X, Hu D H, Liu C X, Yang J, Li Y, Huang Y Q, Feng Y Q, Gong H Y, Li Y, Fang G, Tang H R, Li Y S.2015. Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J Exp Bot, 66(3): 973-987. |
| [44] | Wellburn A R.1994. The spectral determination of chlorophyll a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution.J Plant Physiol, 144(3): 307-313. |
| [45] | Williams B, Dickman M.2008. Plant programmed cell death: Can’t live with it; Can’t live without it.Mol Plant Pathol, 9(4): 531-544. |
| [46] | Wu C J, Bordeos A, Madamba M R S, Baraoidan M, Ramos M, Wang G L, Leach J E, Leung H.2008. Rice lesion mimic mutants with enhanced resistance to diseases.Mol Genet Genom, 279(6): 605-619. |
| [47] | Wu J L, Wu C J, Lei C L, Baraoidan M, Bordeos A, Madamba M R S, Ramos-Pamplona M, Mauleon R, Portugal A, Ulat V J, Bruskiewich R, Wang G L, Leach J, Khush G, Leung H.2005. Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol Biol, 59(1): 85-97. |
| [48] | Xu X, Chen Z, Shi Y F, Wang H M, He Y, Shi L, Chen T, Wu J L, Zhang X B.2018. Functional inactivation of OsGCNT induces enhanced disease resistance to Xanthomonas oryzae pv. oryzae in rice. BMC Plant Biol, 18: 264. |
| [49] | Yamanouchi U, Yano M, Lin H X, Ashikari M, Yamada K.2002. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci USA, 99(11): 7530-7535. |
| [50] | Yin Z C, Chen J, Zeng L R, Goh M, Leung H, Khush G S, Wang G L.2000. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight.Mol Plant Microbe Interact, 13(8): 869-876. |
| [51] | Yuan Y X, Zhong S H, Li Q, Zhu Z R, Lou Y G, Wang L Y, Wang J J, Wang M Y, Li Q L, Yang D L, He Z H.2007. Functional analysis of riceNPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol J, 5(2): 313-324. |
| [52] | Zeng L R, Qu S H, Bordeos A, Yang C W, Baraoidan M, Yan H Y, Xie Q, Nahm B H, Leung H, Wang G L.2004. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell, 16(10): 2795-2808. |
| [53] | Zhang X B, Feng B H, Wang H M, Xu X, Shi Y F, He Y, Chen Z, Sathe A P, Shi L, Wu J L.2018. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J Integr Plant Biol, 60(2): 160-172. |
| [54] | Zhou Q Y, Yu Q, Wang Z Q, Pan Y F, Lv W T, Zhu L L, Chen R Z, He G C.2013. Knockdown of GDCH gene reveals reactive oxygen species-induced leaf senescence in rice. Plant Cell Environ, 36(8): 1476-1489. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | XIA Xiaodong, ZHANG Xiaobo, WANG Zhonghao, CHENG Benyi, Sun Huifeng, XU Xia, GONG Junyi, YANG Shihua, WU Jianli, SHI Yongfeng, XU Rugen. Mapping and Functional Analysis of LE Gene in a Lethal Etiolated Rice Mutant at Seedling Stage [J]. Rice Science, 2023, 30(6): 13-. |
| [7] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [8] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [9] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [10] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [11] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [12] | Jiang Changjie, Liang Zhengwei, Xie Xianzhi. Priming for Saline-Alkaline Tolerance in Rice: Current Knowledge and Future Challenges [J]. Rice Science, 2023, 30(5): 417-425. |
| [13] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [14] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [15] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||