
Rice Science ›› 2022, Vol. 29 ›› Issue (4): 363-374.DOI: 10.1016/j.rsci.2021.11.006
• Research Paper • Previous Articles Next Articles
Wu Zhongling1,#, Qiu Jiehua2,#, Shi Huanbin2, Lin Chuyu1, Yue Jiangnan1, Liu Zhiquan2, Xie Wei1, Naweed I. Naqvi3, Kou Yanjun2( ), Tao Zeng1(
), Tao Zeng1( )
)
Received:2021-08-20
Accepted:2021-11-23
Online:2022-07-28
Published:2022-06-01
Contact:
Kou Yanjun, Tao Zeng
About author:First author contact:#These authors contributed equally to this work
Wu Zhongling, Qiu Jiehua, Shi Huanbin, Lin Chuyu, Yue Jiangnan, Liu Zhiquan, Xie Wei, Naweed I. Naqvi, Kou Yanjun, Tao Zeng. Polycomb Repressive Complex 2-Mediated H3K27 Trimethylation Is Required for Pathogenicity in Magnaporthe oryzae[J]. Rice Science, 2022, 29(4): 363-374.
Add to citation manager EndNote|Ris|BibTeX
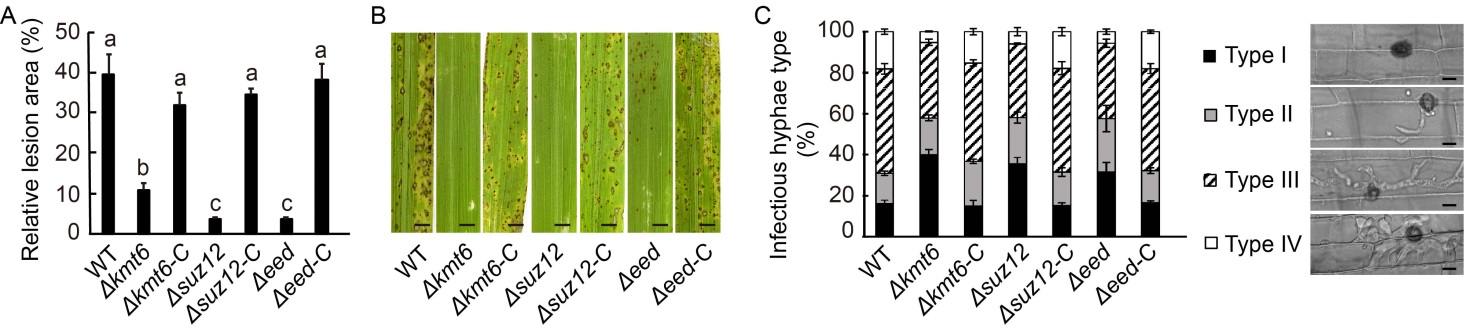
Fig. 1. Polycomb repressive complex 2 (PRC2) is required for fungal pathogenicity in Magnaporthe oryzae. A, Relative lesion area of indicated strains. The relative lesion area was quantified by an ImageJ software. Δkmt6, Δsuz12 and Δeed are deletion mutants from the wild type (WT) strain. Δkmt6-C, Δsuz12-C and Δeed-C are their complementary strains, respectively. B, Blast infection assay using 21-day-old rice seedlings (CO39). Deletion of Kmt6, Eed and Suz12 impaired the pathogenicity of M. oryzae. Scale bars, 2 mm. C, Observation and statistical analysis of invasive hypha growth in rice sheath cells at 40 h post-inoculation. Four types of invasive hyphae: no penetration (Type I), penetration with primary hyphae (Type II), with differentiated secondary invasive hyphae (Type III), and invasive hyphae spreading into neighbouring cells (Type IV), were quantified. Data represent Mean ± SD of three independent repeats, with n = 300 appressoria per analysis. Scale bars, 5 μm.
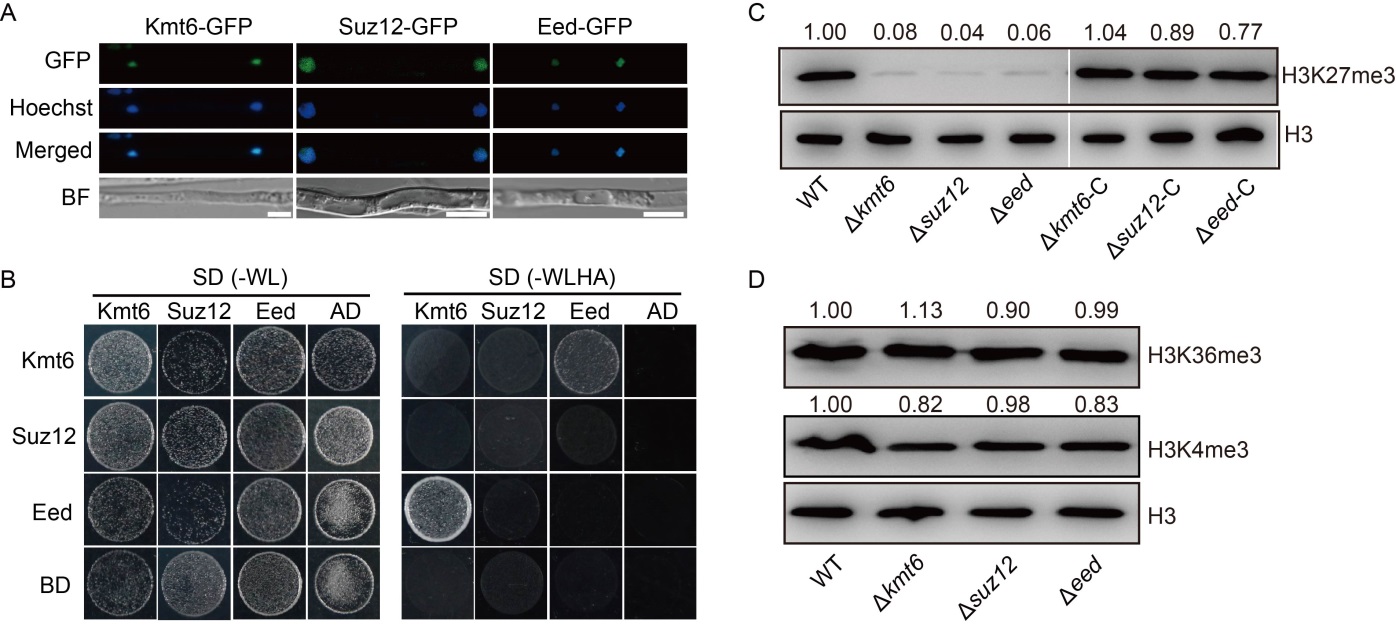
Fig. 2. Core subunits of Polycomb repressive complex 2 (PRC2) are indispensable for H3K27me3 modification in Magnaporthe oryzae. A, Confocal microscopy image for subcellular localization of PRC2 subunits fused with green fluorescent protein (GFP). The GFP signal co-localized with Hoechst (10 μg/mL) stained nuclei. BF, Blight field. Scale bars, 5 μm. B, Yeast two-hybrid assay of PRC2 components Kmt6, Eed and Suz12. The bait and prey plasmids were co-transformed into yeast strain Y2Hgold, respectively. Then, the transformants were grown on basal medium SD (-WL, without tryptophan and leucine) and selective medium SD (-WLHA, without tryptophan, leucine, histidine and adenine). The empty plasmids pGADT7 (AD) and pGBKT7 (BD) were used as controls. C, Levels of histone H3 and H3K27me3 in wild type (WT), ∆kmt6, ∆eed and ∆suz12 strains and their complementary strains (∆kmt6-C, ∆eed-C and ∆suz12-C) on total histones were measured by Western blotting. The relative intensity abundance was measured and calculated. Two repeated biological experiments were conducted with similar results. D, Levels of histone H3, H3K4me3 and H3K36me3 in WT, ∆kmt6, ∆eed and ∆suz12 strains were measured by Western blotting. The relative intensity abundances were measured and calculated. Two repeated biological experiments were conducted with similar results.
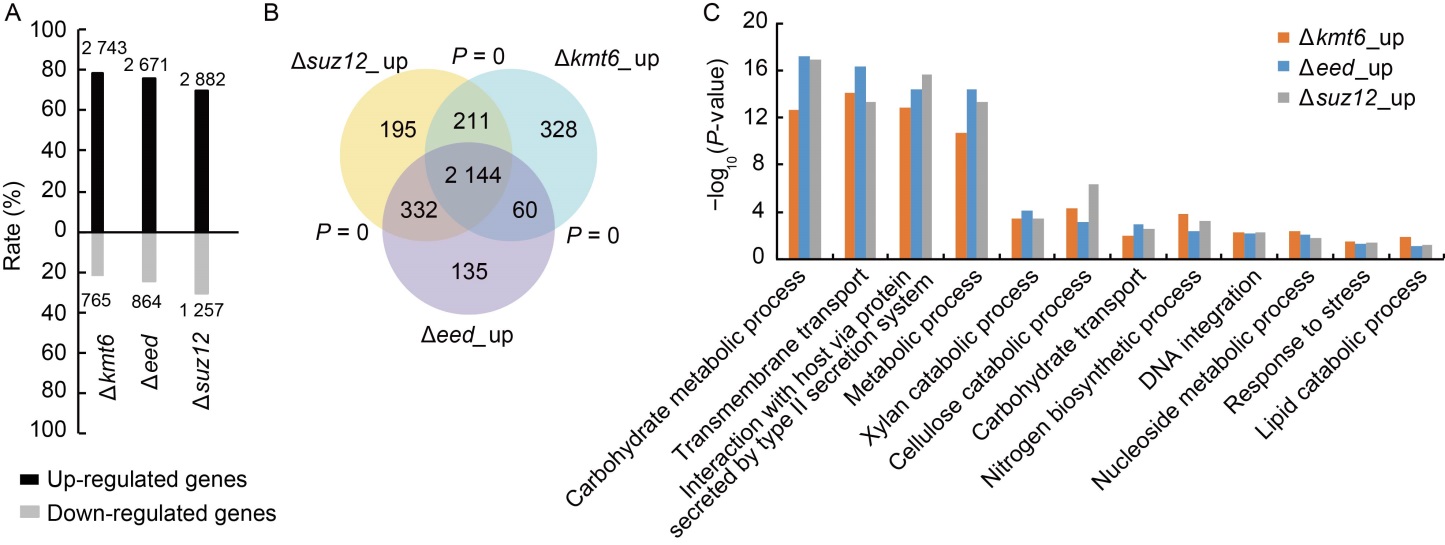
Fig. 3. Core subunits of Polycomb repressive complex 2 (PRC2) regulate genome-wide gene expression in Magnaporthe oryzae. A, Summary of up- and down-regulated genes in different PRC2 deletion mutants with RNA-seq analysis. The numbers at the top or bottom of the bars are the numbers of differential expressed genes in the mutants. The Y-axis is the percentage of up- and down-regulated genes in all the differentially expressed genes. Data were obtained from three independent biological=repeats. B, Venn diagram showing statistically significant overlaps among gene sets of ∆kmt6_up, ∆suz12_up and ∆eed_up. P value for overlapping between gene sets was obtained by the Fisher’s exact test. C, Gene Ontology (GO) analysis of ∆kmt6_up, ∆suz12_up and ∆eed_up genes.
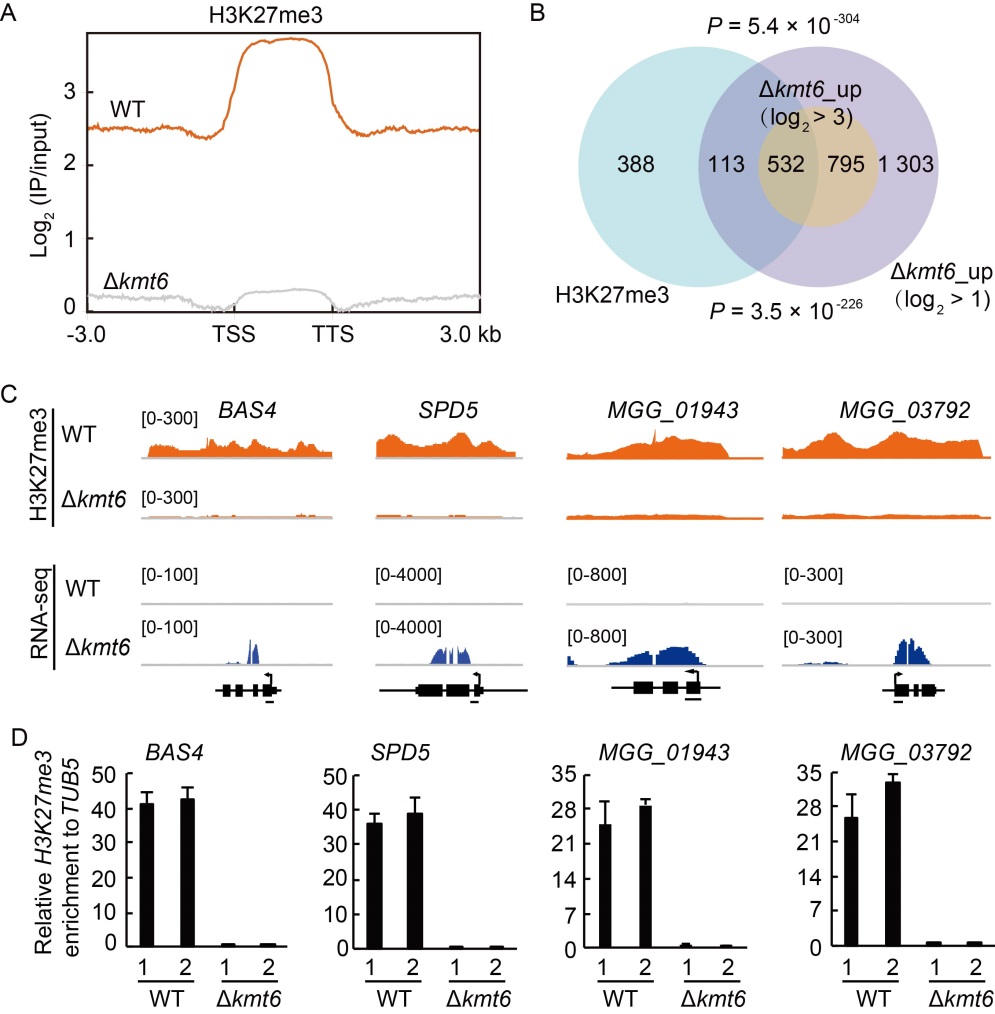
Fig. 4. H3K27me3 occupancy is highly associated with de-repression of target genes in Polycomb repressive complex 2 (PRC2) deletion mutants. A, Average H3K27me3 occupancy within 3-kb genomic regions flanking the summit of H3K27me3 peaks in wild type (WT) and ∆kmt6 strains. TSS, Transcription start site; TTS, Transcription termination site. B, Venn diagram showing statistically significant overlaps between gene sets of H3K27me3-occupied genes and ∆kmt6_up with thresholds of log2 > 1 and log2 > 3 respectively. P values with Fisher’s exact test for overlapping between gene sets were labeled. C, Genome browser views of H3K27me3 occupancy from chromatin immune- precipitation assay followed by high- throughout sequencing (ChIP-seq) and expression pattern from RNA-seq of selected genes. The number areas were reads per million (RPM). D, ChIP-qPCR verified the enrichment of H3K27me3 at the chromatin of selected genes. The examined regions were shown with black line at the bottom of gene model. The relative enrichments were calculated by relative quantitation from two biological repeats, which was standardized with an internal control TUB5, then compared with that of WT. The numbers ‘1’ and ‘2’ indicated two independent repeats. Values are Mean ± SD of three technical repeats.
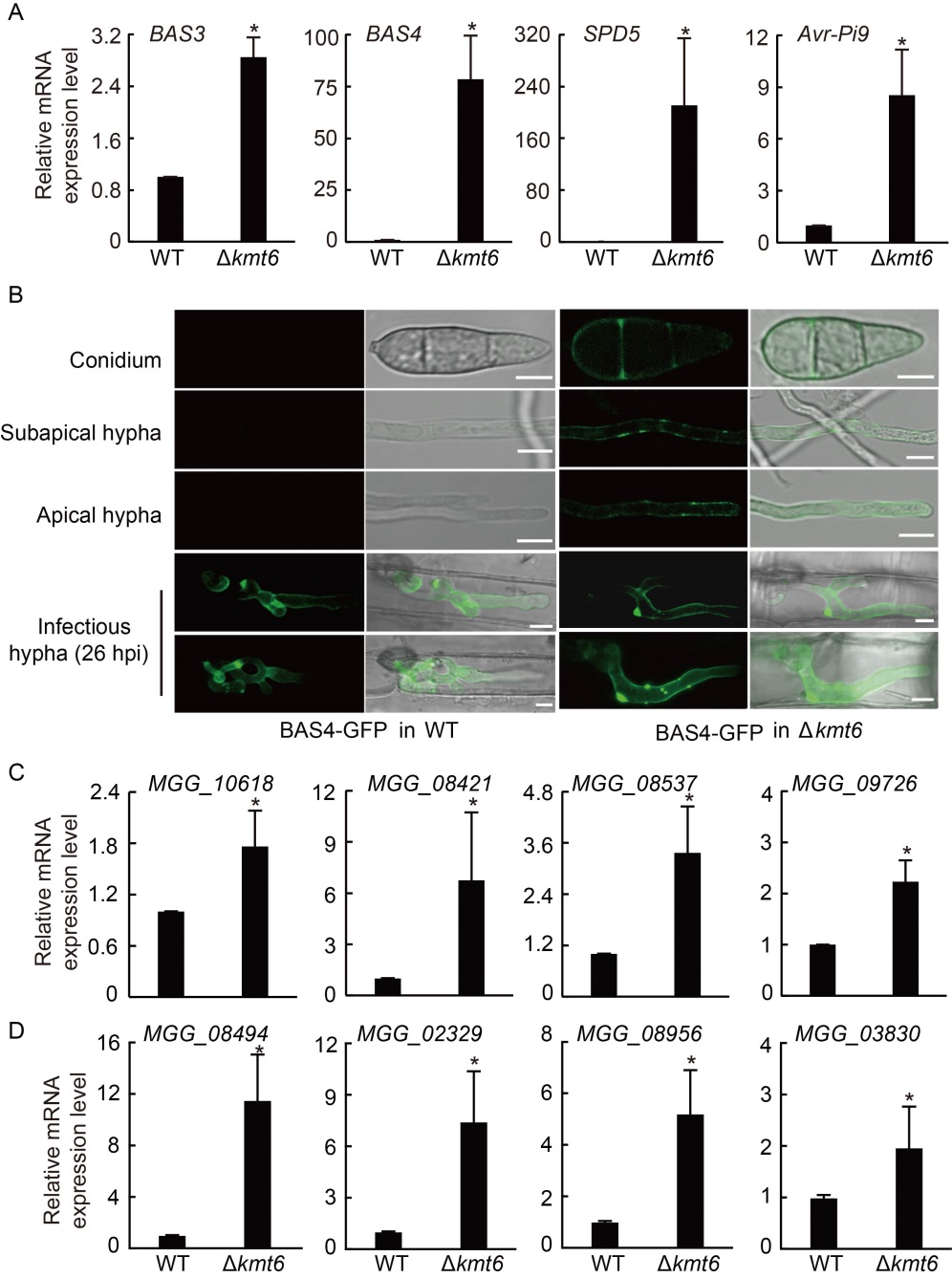
Fig. 5. Expression levels of genes which encode effector, extracellular enzyme and cytochrome P450 were up-regulated in Polycomb repressive complex 2 (PRC2) deletion mutants. A, Relative expression levels of effectors were checked in wild type (WT) and Δkmt6 strains. The strains were cultured in liquid complete medium (CM) at 28 ºC for 2 d. Tubulin was used as an internal reference. B, Confocal microscopy image of Bas4- GFP at the vegetative and in planta growth stages of WT and ∆kmt6 strains. In WT, Bas4-GFP fluorescence was only shown at the in planta growth stage, while in ∆kmt6, Bas4-GFP fluorescence can be detected in the conidia, apical hyphae, subapical hyphae and invasive hyphae. GFP, Green fluorescent protein; hpi, Hours post-inoculation. Scale bars, 5 μm. C and D, Relative expression levels of pathogenic genes that encode extracellular enzyme (C) and cytochrome P450 (D) were checked in WT and Δkmt6 strains. Tubulin was used as an internal reference. The strains were cultured in liquid CM at 28 ºC for 2 d. Values are Mean ± SD of three biological repeats. *, Significant differences at P < 0.05 between the deletion mutant and WT strains by the Student’s t-test.
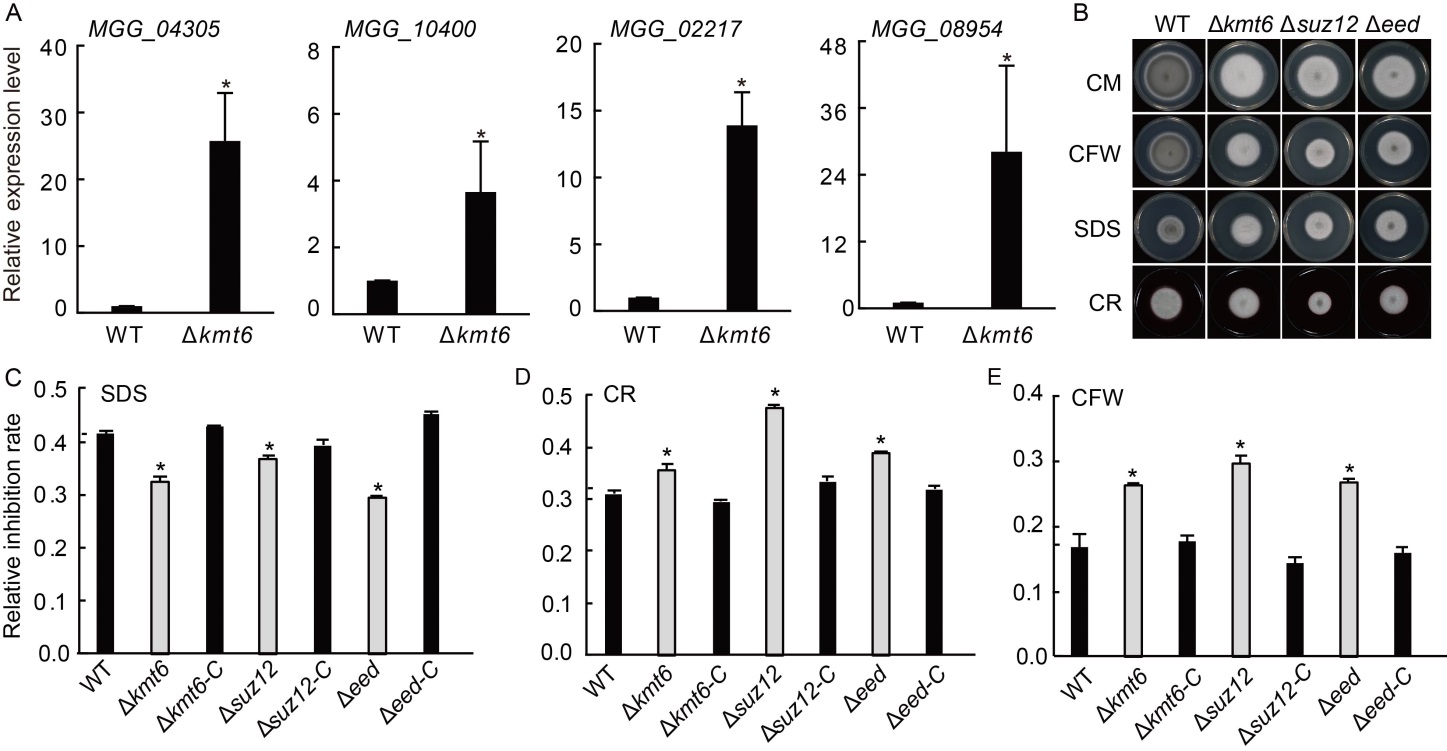
Fig. 6. Polycomb repressive complex 2 (PRC2) is required for stress response in Magnaporthe oryzae. A, Relative expression levels of cell wall related genes. Values are Mean ± SD of three biological repeats. *, Significant differences at P < 0.05 between the deletion mutant and wild type (WT) strains by the Student’s t-test. The strains were cultured in liquid CM at 28 ºC for 2 d. Tubulin was used as an internal reference. B, Radical growths of WT, ∆kmt6, ∆eed, ∆suz12 and their complementary strains (∆kmt6-C, ∆eed-C and ∆suz12-C). Colonies of indicated strains were grown on CM supplemented with different chemicals for 7 d. C?E, Statistical analysis of colony diameters of tested strains on CM supplemented with 0.005% SDS (C), 0.1 mg/mL CR (D), and 0.05 mg/mL CFW (E) for 7 d. Values are Mean ± SD of three independent repeats. *, Significant differences at P < 0.05 between the deletion mutant and WT strains by the Student’s t-test. CM, Complete medium; SDS, Sodium dodecyl sulphate; CR, Congo red; CFW, Calcofluor white.
| [1] | Blackledge N P, Rose N R, Klose R J. 2015. Targeting Polycomb systems to regulate gene expression: Modifications to a complex story. Nat Rev Mol Cell Biol, 16: 643-649. |
| [2] | Cavalli G, Heard E. 2019. Advances in epigenetics link genetics to the environment and disease. Nature, 571: 489-499. |
| [3] | Chadha S, Sharma M. 2014. Transposable elements as stress adaptive capacitors induce genomic instability in fungal pathogen Magnaporthe oryzae. PLoS One, 9: e94415. |
| [4] | Chujo T, Scott B. 2014. Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte- plant symbiosis. Mol Microbiol, 92: 413-434. |
| [5] | Connolly L R, Smith K M, Freitag M. 2013. The Fusarium graminearum histone H3 K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. PLoS Genet, 9: e1003916. |
| [6] | Ding S L, Liu W D, Iliuk A, Ribot C, Vallet J, Tao A, Wang Y, Lebrun M H, Xu J R. 2010. The Tig1 histone deacetylase complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae. Plant Cell, 22: 2495-2508. |
| [7] | Dong Y H, Li Y, Zhao M M, Jing M F, Liu X Y, Liu M X, Guo X X, Zhang X, Chen Y, Liu Y F, Liu Y H, Ye W W, Zhang H F, Wang Y C, Zheng X B, Wang P, Zhang Z G. 2015. Global genome and transcriptome analyses of Magnaporthe oryzae epidemic isolate 98-06 uncover novel effectors and pathogenicity- related genes, revealing gene gain and lose dynamics in genome evolution. PLoS Pathog, 11: e1004801. |
| [8] | Dong Y H, Li Y, Qi Z Q, Zheng X B, Zhang Z G. 2016. Genome plasticity in filamentous plant pathogens contributes to the emergence of novel effectors and their cellular processes in the host. Curr Genet, 62: 47-51. |
| [9] | Dumesic P A, Homer C M, Moresco J J, Pack L R, Shanle E K, Coyle S M, Strahl B D, Fujimori D G, Yates III J R, Madhani H D. 2015. Product binding enforces the genomic specificity of a yeast Polycomb repressive complex. Cell, 160: 204-218. |
| [10] | Feng W Z, Yin Z Y, Wu H W, Liu P, Liu X Y, Liu M X, Yu R, Gao C Y, Zhang H F, Zheng X B, Wang P, Zhang Z G. 2021. Balancing of the mitotic exit network and cell wall integrity signaling governs the development and pathogenicity in Magnaporthe oryzae. PLoS Pathog, 17: e1009080. |
| [11] | Fouché S, Plissonneau C, Croll D. 2018. The birth and death of effectors in rapidly evolving filamentous pathogen genomes. Curr Opin Microbiol, 46: 34-42. |
| [12] | Galazka J M, Freitag M. 2014. Variability of chromosome structure in pathogenic fungi: Of ‘ends and odds’. Curr Opin Microbiol, 20: 19-26. |
| [13] | He M, Xu Y P, Chen J H, Luo Y, Lv Y, Su J, Kershaw M J, Li W T, Wang J, Yin J J, Zhu X B, Liu X H, Chern M, Ma B T, Wang J C, Qin P, Chen W L, Wang Y P, Wang W M, Ren Z L, Wu X J, Li P, Li S G, Peng Y L, Lin F C, Talbot N J, Chen X W. 2018. MoSnt2-dependent deacetylation of histone H3 mediates MoTor- dependent autophagy and plant infection by the rice blast fungus Magnaporthe oryzae. Autophagy, 14: 1543-1561. |
| [14] | Jamieson K, Rountree M R, Lewis Z A, Stajich J E, Selker E U. 2013. Regional control of histone H3 lysine 27 methylation in Neurospora. Proc Natl Acad Sci USA, 110: 6027-6032. |
| [15] | Kassis J A, Brown J L. 2013. Polycomb group response elements in Drosophila and vertebrates. Adv Genet, 81: 83-118. |
| [16] | Kawahara Y, Oono Y, Kanamori H, Matsumoto T, Itoh T, Minami E. 2012. Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS One, 7: e49423. |
| [17] | Khang C H, Berruyer R, Giraldo M C, Kankanala P, Park S Y, Czymmek K, Kang S, Valent B. 2010. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell, 22: 1388-1403. |
| [18] | Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S L. 2013. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol, 14: R36. |
| [19] | Kou Y J, Tan Y H, Ramanujam R, Naqvi N I. 2017. Structure- function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol, 214: 330-342. |
| [20] |
Langmead B, Salzberg S L. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods, 9: 357-359.
PMID |
| [21] | Langmead B, Trapnell C, Pop M, Salzberg S L. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol, 10: R25. |
| [22] |
Lanzuolo C, Orlando V. 2012. Memories from the polycomb group proteins. Annu Rev Genet, 46: 561-589.
PMID |
| [23] | Lee J, Lee J J, Jeon J. 2019. A histone deacetylase, MoHOS2 regulates asexual development and virulence in the rice blast fungus. J Microbiol, 57: 1115-1125. |
| [24] | Li Z C, Fu X, Wang Y Z, Liu R Y, He Y H. 2018. Polycomb- mediated gene silencing by the BAH-EMF1 complex in plants. Nat Genet, 50: 1254-1261. |
| [25] | Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature, 469: 343-349. |
| [26] | Mathioni S M, Patel N, Riddick B, Sweigard J A, Czymmek K J, Caplan J L, Kunjeti S G, Kunjeti S, Raman V, Hillman B I, Kobayashi D Y, Donofrio N M. 2013. Transcriptomics of the rice blast fungus Magnaporthe oryzae in response to the bacterial antagonist Lysobacter enzymogenes reveals candidate fungal defense response genes. PLoS One, 8: e76487. |
| [27] |
Mosquera G, Giraldo M C, Khang C H, Coughlan S, Valent B. 2009. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell, 21: 1273-1290.
PMID |
| [28] | Netea M G, Joosten L A B, Latz E, Mills K H G, Natoli G, Stunnenberg H G, O'Neill L A, Xavier R J. 2016. Trained immunity: A program of innate immune memory in health and disease. Science, 352: aaf1098. |
| [29] | Oh Y, Donofrio N, Pan H Q, Coughlan S, Brown D E, Meng S W, Mitchell T, Dean R A. 2008. Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol, 9: R85. |
| [30] | Pham K T M, Inoue Y, Vu B V, Nguyen H H, Nakayashiki T, Ikeda K I, Nakayashiki H. 2015. MoSET1 (histone H3K4 methyltransferase in Magnaporthe oryzae) regulates global gene expression during infection-related morphogenesis. PLoS Genet, 11: e1005385. |
| [31] | Qian B, Liu X Y, Ye Z Y, Zhou Q K, Liu P, Yin Z Y, Wang W H, Zheng X B, Zhang H F, Zhang Z G. 2021. Phosphatase- associated protein MoTip41 interacts with the phosphatase MoPpe1 to mediate crosstalk between TOR and cell wall integrity signalling during infection by the rice blast fungus Magnaporthe oryzae. Environ Microbiol, 23: 791-809. |
| [32] | Ridenour J B, Möller M, Freitag M. 2020. Polycomb repression without bristles: Facultative heterochromatin and genome stability in fungi. Genes, 11(6): 638. |
| [33] |
Robinson J T, Thorvaldsdóttir H, Winckler W, Guttman M, Lander E S, Getz G, Mesirov J P. 2011. Integrative genomics viewer. Nat Biotechnol, 29: 24-26.
PMID |
| [34] |
Sánchez-Vallet A, Fouché S, Fudal I, Hartmann F E, Soyer J L, Tellier A, Croll D. 2018. The genome biology of effector gene evolution in filamentous plant pathogens. Annu Rev Phytopathol, 56: 21-40.
PMID |
| [35] |
Schuettengruber B, Bourbon H M, di Croce L, Cavalli G. 2017. Genome regulation by polycomb and trithorax: 70 years and counting. Cell, 171: 34-57.
PMID |
| [36] | Sharpee W, Oh Y, Yi M, Franck W, Eyre A, Okagaki L H, Valent B, Dean R A. 2017. Identification and characterization of suppressors of plant cell death (SPD) effectors from Magnaporthe oryzae. Mol Plant Pathol, 18: 850-863. |
| [37] | Tao Z, Shen L S, Gu X F, Wang Y Z, Yu H, He Y H. 2017. Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature, 551: 124-128. |
| [38] |
Trapnell C, Williams B A, Pertea G, Mortazavi A, Kwan G, van Baren M J, Salzberg S L, Wold B J, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol, 28: 511-515.
PMID |
| [39] | Villalba F, Collemare J, Landraud P, Lambou K, Brozek V, Cirer B, Morin D, Bruel C, Beffa R, Lebrun M H. 2008. Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet Biol, 45: 68-75. |
| [40] | Wang L Y, Chen H, Li J J, Shu H D, Zhang X X, Wang Y C, Tyler B M, Dong S M. 2020. Effector gene silencing mediated by histone methylation underpins host adaptation in an oomycete plant pathogen. Nucleic Acids Res, 48: 1790-1799. |
| [41] | Wiles E T, Selker E U. 2017. H3K27 methylation: A promiscuous repressive chromatin mark. Curr Opin Genet Dev, 43: 31-37. |
| [42] | Wiles E T, McNaught K J, Kaur G, Selker J M L, Ormsby T, Aravind L, Selker E U. 2020. Evolutionarily ancient BAH-PHD protein mediates Polycomb silencing. Proc Natl Acad Sci USA, 117: 11614-11623. |
| [43] |
Xiao J, Jin R, Yu X, Shen M, Wagner J D, Pai A, Song C, Zhuang M, Klasfeld S, He C S, Santos A M, Helliwell C, Pruneda-Paz J L, Kay S A, Lin X W, Cui S J, Garcia M F, Clarenz O, Goodrich J, Zhang X Y, Austin R S, Bonasio R, Wagner D. 2017. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat Genet, 49: 1546-1552.
PMID |
| [44] | Yang Z L, Qian S M, Scheid R N, Lu L, Chen X S, Liu R, Du X, Lv X C, Boersma M D, Scalf M, Smith L M, Denu J M, Du J M, Zhong X H. 2018. EBS is a bivalent histone reader that regulates floral phase transition in Arabidopsis. Nat Genet, 50: 1247-1253. |
| [45] | Zhang S L, Liang M L, Naqvi N I, Lin C X, Qian W Q, Zhang L H, Deng Y Z. 2017. Phototrophy and starvation-based induction of autophagy upon removal of Gcn5-catalyzed acetylation of Atg7 in Magnaporthe oryzae. Autophagy, 13: 1318-1330. |
| [46] | Zhang W, Huang J, Cook D E. 2021. Histone modification dynamics at H3K27 are associated with altered transcription of in planta induced genes in Magnaporthe oryzae. PLoS Genet, 17: e1009376. |
| [47] | Zhang Y, Liu T, Meyer C A, Eeckhoute J, Johnson D S, Bernstein B E, Nusbaum C, Myers R M, Brown M, Li W, Liu X S. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol, 9: R137. |
| [1] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [2] | Liu Yueran, Qu Jinsong, Wang Yufu, Yin Weixiao, Luo Chaoxi. bZIP Transcription Factor UvATF21 Mediates Vegetative Growth, Conidiation, Stress Tolerance and Is Required for Full Virulence of Rice False Smut Fungus Ustilaginoidea virens [J]. Rice Science, 2023, 30(1): 50-57. |
| [3] | Suhas Gorakh Karkute, Vishesh Kumar, Mohd Tasleem, Dwijesh Chandra Mishra, Krishna Kumar Chaturvedi, Anil Rai, Amitha Mithra Sevanthi, Kishor Gaikwad, Tilak Raj Sharma, Amolkumar U. Solanke. Genome-Wide Analysis of von Willebrand Factor A Gene Family in Rice for Its Role in Imparting Biotic Stress Resistance with Emphasis on Rice Blast Disease [J]. Rice Science, 2022, 29(4): 375-384. |
| [4] | Zhou Ying, Wan Tao, Yuan Bin, Lei Fang, Chen Meijuan, Wang Qiong, Huang Ping, Kou Shuyan, Qiu Wenxiu, Liu Li. Improving Rice Blast Resistance by Mining Broad-Spectrum Resistance Genes at Pik Locus [J]. Rice Science, 2022, 29(2): 133-142. |
| [5] | Junjie Yu, Mina Yu, Tianqiao Song, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Xiaole Yin, Dong Liang, Yongfeng Liu. UvSMEK1, a Suppressor of MEK Null, Regulates Pathogenicity, Conidiation and Conidial Germination in Rice False Smut Fungus Ustilaginoidea virens [J]. Rice Science, 2021, 28(5): 457-465. |
| [6] | Junhua Lu, Xuemei Yang, Jinfeng Chen, Tingting Li, Zijin Hu, Ying Xie, Jinlu Li, Jiqun Zhao, Mei Pu, Hui Feng, Jing Fan, Yanyan Huang, Jiwei Zhang, Wenming Wang, Yan Li. Osa-miR439 Negatively Regulates Rice Immunity Against Magnaporthe oryzae [J]. Rice Science, 2021, 28(2): 156-165. |
| [7] | Yanchang Luo, Tingchen Ma, Teo Joanne, Zhixiang Luo, Zefu Li, Jianbo Yang, Zhongchao Yin. Marker-Assisted Breeding of Thermo-Sensitive Genic Male Sterile Line 1892S for Disease Resistance and Submergence Tolerance [J]. Rice Science, 2021, 28(1): 89-98. |
| [8] | Ning Xiao, Yunyu Wu, Aihong Li. Strategy for Use of Rice Blast Resistance Genes in Rice Molecular Breeding [J]. Rice Science, 2020, 27(4): 263-277. |
| [9] | B. ANGELES-SHIM Rosalyn, P. REYES Vincent, M. del VALLE Marilyn, S. LAPIS Ruby, SHIM Junghyun, SUNOHARA Hidehiko, K. JENA Kshirod, ASHIKARI Motoyuki, DOI Kazuyuki. Marker-Assisted Introgression of Quantitative Resistance Gene pi21 Confers Broad Spectrum Resistance to Rice Blast [J]. Rice Science, 2020, 27(2): 113-123. |
| [10] | Challagulla Vineela, Bhattarai Surya, J. Midmore David. In-vitro vs in-vivo Inoculation: Screening for Resistance of Australian Rice Genotypes Against Blast Fungus [J]. Rice Science, 2015, 22(3): 132-137. |
| [11] | WANG Jiao-yu1, WANG Xiao-yan1, 2, LI Ling1, ZHANG Xin1, WANG Yan-li1, CHAI Rong-yao1, SUN Guo-chang1. Pathogenicity of Rice Blast Fungus Magnaporthe oryzae on Brachypodium distachyon [J]. RICE SCIENCE, 2012, 19(3): 252-258. |
| [12] | Y J P K MITHRASENA1, W S S WIJESUNDERA2, R L C WIJESUNDERA3, D C WIMALASIRI2, R P N PRIYANTHI2. Pathogenic and Genetic Diversity of Magneporthe oryzae Populations from Sri Lanka [J]. RICE SCIENCE, 2012, 19(3): 241-246. |
| [13] | CHEN De-xi, CHEN Xue-wei, LEI Cai-lin, MA Bing-tian, WANG Yu-ping, LI Shi-gui. Rice Blast Resistance of Transgenic Rice Plants with Pi-d2 Gene [J]. RICE SCIENCE, 2010, 17(3): 179-184 . |
| [14] | CHEN De-xi, CHEN Xue-wei, MA Bing-tian, WANG Yu-ping, ZHU Li-huang, LI Shi-gui, . Genetic Transformation of Rice with Pi-d2 Gene Enhances Resistance to Rice Blast Fungus Magnaporthe oryzae [J]. RICE SCIENCE, 2010, 17(1): 19-27 . |
| [15] | YANG Jian-yuan, CHEN Shen, ZENG Lie-xian, LI Yi-long, CHEN Zhen, LI Chuan-ying, ZHU Xiao-yuan. Race Specificity of Major Rice Blast Resistance Genes to Magnaporthe grisea Isolates Collected from indica Rice in Guangdong, China [J]. RICE SCIENCE, 2008, 15(4): 311-318 . |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||