
Rice Science ›› 2022, Vol. 29 ›› Issue (2): 133-142.DOI: 10.1016/j.rsci.2022.01.002
• Research Paper • Previous Articles Next Articles
Zhou Ying1,#( ), Wan Tao1,#, Yuan Bin3, Lei Fang4, Chen Meijuan5, Wang Qiong1, Huang Ping6, Kou Shuyan6, Qiu Wenxiu1(
), Wan Tao1,#, Yuan Bin3, Lei Fang4, Chen Meijuan5, Wang Qiong1, Huang Ping6, Kou Shuyan6, Qiu Wenxiu1( ), Liu Li2(
), Liu Li2( )
)
Received:2021-06-03
Accepted:2021-08-16
Online:2022-03-28
Published:2022-02-09
Contact:
Zhou Ying, Qiu Wenxiu, Liu Li
About author:First author contact:#These authors contributed equally to this work
Zhou Ying, Wan Tao, Yuan Bin, Lei Fang, Chen Meijuan, Wang Qiong, Huang Ping, Kou Shuyan, Qiu Wenxiu, Liu Li. Improving Rice Blast Resistance by Mining Broad-Spectrum Resistance Genes at Pik Locus[J]. Rice Science, 2022, 29(2): 133-142.
Add to citation manager EndNote|Ris|BibTeX
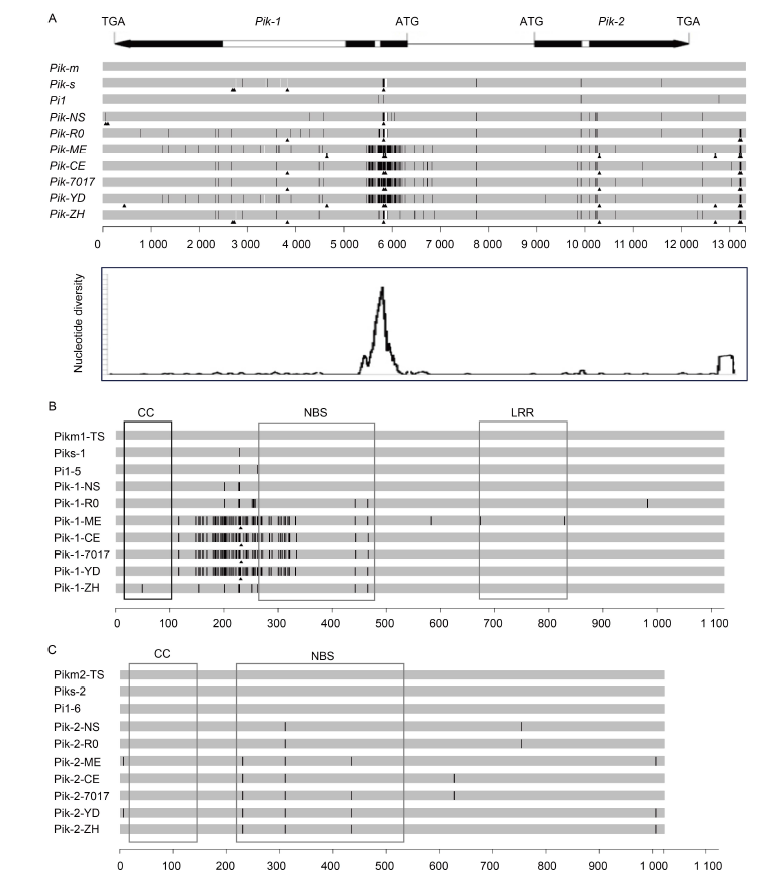
Fig. 1. Schematic maps for sequence alignment of novel Pik alleles, Pik-1 and Pik-2 proteins. Pik alleles, Pik-1 and Pik-2 proteins. A, Schematic diagram of gene structure was shown above, black boxes indicate exons and white boxes indicate introns, and the start codon and the termination codon are labeled with ATG and TGA, respectively. Seven alleles of Pik were isolated from the studied rice accessions and were compared with Pik-m gene as shown in the middle, the unit scale indicates the location of nucleotides. Sliding-window analysis of nucleotide diversity (π) about novel Pik alleles was shown below. B and C, Seven alleles of Pik-1 (B) and Pik-2 proteins (C) were compared with Pik-m protein. The unit scale indicates the location of amino acids. The black line on the bar indicates the amino acid polymorphism compared with the reference sequence. The gap between allele strips indicates deletion, and the size of gap indicates the length of deletion sequence. CC, Coiled coil; NBS, Nucleotide-binding site; LRR, Leucine rich repeat.
| Pik allele | Accession a | No. of accessions carrying the allele | Identity to Pik-m (%) | No. of SNPs | No. of InDels | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Pik-1 | Pik-2 | Total | Pik-1 | Pik-2 | |||||
| Pik-m | Tsuyuake | 13 | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | |
| Pik-s | Khao Phoi | 15 | 99.97 | 4 | 3 | 0 | 0 | 0 | 0 | |
| Pi1 | Tetep | 4 | 99.91 | 12 | 3 | 1 | 5 | 5 | 0 | |
| Pik-NS | Nsicrc 122 | 19 | 99.85 | 23 | 8 | 6 | 7 | 5 | 0 | |
| Pik-R0 | R03138 | 10 | 98.34 | 230 | 20 | 6 | 200 | 6 | 0 | |
| Pik-ME | Meixiangzhan | 1 | 97.17 | 389 | 116 | 9 | 257 | 53 | 0 | |
| Pik-CE | IR65482-4-136-2-2-B | 1 | 97.31 | 373 | 112 | 7 | 246 | 55 | 0 | |
| Pik-7017 | Red Khosha Cerma | 4 | 97.31 | 372 | 105 | 8 | 246 | 49 | 0 | |
| Pik-YD | Yangdao 4038 | 49 | 97.16 | 390 | 113 | 9 | 259 | 62 | 0 | |
| Pik-ZH | Zhouhui 338 | 3 | 98.28 | 232 | 15 | 8 | 201 | 4 | 0 | |
Table 1. Single nucleotide polymorphisms (SNPs) and different alleles of Pik gene in various rice species.
| Pik allele | Accession a | No. of accessions carrying the allele | Identity to Pik-m (%) | No. of SNPs | No. of InDels | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Pik-1 | Pik-2 | Total | Pik-1 | Pik-2 | |||||
| Pik-m | Tsuyuake | 13 | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | |
| Pik-s | Khao Phoi | 15 | 99.97 | 4 | 3 | 0 | 0 | 0 | 0 | |
| Pi1 | Tetep | 4 | 99.91 | 12 | 3 | 1 | 5 | 5 | 0 | |
| Pik-NS | Nsicrc 122 | 19 | 99.85 | 23 | 8 | 6 | 7 | 5 | 0 | |
| Pik-R0 | R03138 | 10 | 98.34 | 230 | 20 | 6 | 200 | 6 | 0 | |
| Pik-ME | Meixiangzhan | 1 | 97.17 | 389 | 116 | 9 | 257 | 53 | 0 | |
| Pik-CE | IR65482-4-136-2-2-B | 1 | 97.31 | 373 | 112 | 7 | 246 | 55 | 0 | |
| Pik-7017 | Red Khosha Cerma | 4 | 97.31 | 372 | 105 | 8 | 246 | 49 | 0 | |
| Pik-YD | Yangdao 4038 | 49 | 97.16 | 390 | 113 | 9 | 259 | 62 | 0 | |
| Pik-ZH | Zhouhui 338 | 3 | 98.28 | 232 | 15 | 8 | 201 | 4 | 0 | |
| Pik allele protein | Accession a | No. of amino acids b | Identity to Pik-m protein (%) | No. of different amino acids | No. of InDels | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pik-1 | Pik-2 | Pik-1 | Pik-2 | Total | Pik-1 | Pik-2 | Total | Pik-1 | Pik-2 | |||||
| Pik-m | Tsuyuake | 1 143 | 1 021 | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | |||
| Pik-s | Khao Phoi | 1 143 | 1 021 | 99.83 | 100.00 | 2 | 2 | 0 | 0 | 0 | 0 | |||
| Pi1 | Tetep | 1 143 | 1 021 | 99.91 | 99.90 | 1 | 1 | 0 | 0 | 0 | 0 | |||
| Pik-NS | Nsicrc 122 | 1 143 | 1 021 | 99.74 | 99.80 | 5 | 3 | 2 | 0 | 0 | 0 | |||
| Pik-R0 | R03138 | 1 143 | 1 021 | 99.13 | 99.80 | 8 | 6 | 2 | 0 | 0 | 0 | |||
| Pik-ME | Meixiangzhan | 1 146 | 1 021 | 94.59 | 99.51 | 67 | 62 | 5 | 3 | 3 | 0 | |||
| Pik-CE | IR65482-4-136-2-2-B | 1 142 | 1 021 | 95.01 | 99.71 | 60 | 57 | 3 | 1 | 1 | 0 | |||
| Pik-7017 | Red Khosha Cerma | 1 142 | 1 021 | 95.10 | 99.61 | 60 | 56 | 4 | 1 | 1 | 0 | |||
| Pik-YD | Yangdao 4038 | 1 144 | 1 021 | 94.85 | 99.51 | 64 | 59 | 5 | 1 | 1 | 0 | |||
| Pik-ZH | Zhouhui 338 | 1 143 | 1 021 | 99.21 | 99.61 | 13 | 9 | 4 | 0 | 0 | 0 | |||
Table 2. Summary of difference in each allele of Pik protein.
| Pik allele protein | Accession a | No. of amino acids b | Identity to Pik-m protein (%) | No. of different amino acids | No. of InDels | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pik-1 | Pik-2 | Pik-1 | Pik-2 | Total | Pik-1 | Pik-2 | Total | Pik-1 | Pik-2 | |||||
| Pik-m | Tsuyuake | 1 143 | 1 021 | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | |||
| Pik-s | Khao Phoi | 1 143 | 1 021 | 99.83 | 100.00 | 2 | 2 | 0 | 0 | 0 | 0 | |||
| Pi1 | Tetep | 1 143 | 1 021 | 99.91 | 99.90 | 1 | 1 | 0 | 0 | 0 | 0 | |||
| Pik-NS | Nsicrc 122 | 1 143 | 1 021 | 99.74 | 99.80 | 5 | 3 | 2 | 0 | 0 | 0 | |||
| Pik-R0 | R03138 | 1 143 | 1 021 | 99.13 | 99.80 | 8 | 6 | 2 | 0 | 0 | 0 | |||
| Pik-ME | Meixiangzhan | 1 146 | 1 021 | 94.59 | 99.51 | 67 | 62 | 5 | 3 | 3 | 0 | |||
| Pik-CE | IR65482-4-136-2-2-B | 1 142 | 1 021 | 95.01 | 99.71 | 60 | 57 | 3 | 1 | 1 | 0 | |||
| Pik-7017 | Red Khosha Cerma | 1 142 | 1 021 | 95.10 | 99.61 | 60 | 56 | 4 | 1 | 1 | 0 | |||
| Pik-YD | Yangdao 4038 | 1 144 | 1 021 | 94.85 | 99.51 | 64 | 59 | 5 | 1 | 1 | 0 | |||
| Pik-ZH | Zhouhui 338 | 1 143 | 1 021 | 99.21 | 99.61 | 13 | 9 | 4 | 0 | 0 | 0 | |||
| Gene | Accession a | Enshi | Yichang | Jiamusi | ||||
|---|---|---|---|---|---|---|---|---|
| 2018 | 2019 | 2018 | 2019 | 2018 | ||||
| CK1 | Lijiangxintuanheigu | S | S | HS | S | HS | ||
| CK2 | Jin 23B | HS | HS | HS | HS | S | ||
| Pik-m | R107 | HS | HS | HS | HS | MR | ||
| Pik-s | Jiahua 1 | R | HR | HR | HR | MR | ||
| Pi1 | Tetep | HR | HR | HR | HR | S | ||
| Pik-NS | Nsicrc 122 | HR | HR | HR | HR | MR | ||
| Pik-R0 | R03138 | HR | HR | HR | HR | MR | ||
| Pik-ME | Meixiangzhan | HR | HR | MR | MR | MR | ||
| Pik-CE | IR65482-4-136-2-2-B | S | MR | HR | MR | MR | ||
| Pik-7017 | Red Khosha Cerma | MS | HR | MS | MR | MS | ||
| Pik-YD | Yangdao 4038 | R | HR | HR | HR | R | ||
| Pik-ZH | Zhouhui 338 | HS | HS | HS | HS | MR | ||
Table 3. Disease responses of cloned genes and Pik allele honor plants in field text.
| Gene | Accession a | Enshi | Yichang | Jiamusi | ||||
|---|---|---|---|---|---|---|---|---|
| 2018 | 2019 | 2018 | 2019 | 2018 | ||||
| CK1 | Lijiangxintuanheigu | S | S | HS | S | HS | ||
| CK2 | Jin 23B | HS | HS | HS | HS | S | ||
| Pik-m | R107 | HS | HS | HS | HS | MR | ||
| Pik-s | Jiahua 1 | R | HR | HR | HR | MR | ||
| Pi1 | Tetep | HR | HR | HR | HR | S | ||
| Pik-NS | Nsicrc 122 | HR | HR | HR | HR | MR | ||
| Pik-R0 | R03138 | HR | HR | HR | HR | MR | ||
| Pik-ME | Meixiangzhan | HR | HR | MR | MR | MR | ||
| Pik-CE | IR65482-4-136-2-2-B | S | MR | HR | MR | MR | ||
| Pik-7017 | Red Khosha Cerma | MS | HR | MS | MR | MS | ||
| Pik-YD | Yangdao 4038 | R | HR | HR | HR | R | ||
| Pik-ZH | Zhouhui 338 | HS | HS | HS | HS | MR | ||
| Gene | Generation | Accession a | Resistant rate (%) b | Disease score c |
|---|---|---|---|---|
| CK1 | Parent | Lijiangxintuanheigu | 0.00 | 8.94 ± 0.36 |
| CK2 | Parent | Jin 23B | 3.23 | 7.90 ± 1.54 |
| CK3 | Parent | Kongyu 131 | 0.00 | 7.45 ± 1.84 |
| Pik-m | Parent | Tsuyuake | 77.42 | 3.03 ± 2.59 |
| Pik-s | Parent | Jiahua 1 | 100.00 | 1.71 ± 1.03 |
| Pi1 | Parent | Tetep | 87.10 | 2.55 ± 2.22 |
| Pik-NS | Parent | AMOL3 | 25.81 | 5.90 ± 2.77 |
| Pik-R0 | Parent | R03138 | 96.77 | 0.97 ± 1.14 |
| Pik-ME | Parent | Meixiangzhan | 80.65 | 2.35 ± 2.09 |
| Pik-CE | Parent | IR65482-4-136-2-2-B | 77.42 | 2.48 ± 2.42 |
| Pik-7017 | Parent | IR70175-22-1-1-2-2 | 100.00 | 0.52 ± 0.51 |
| Pik-YD | Parent | Yangdao 4038 | 77.42 | 2.65 ± 2.88 |
| Pik-ZH | Parent | Zhouhui 338 | 96.77 | 4.94 ± 2.10 |
| Pik-m | BC3F2 | Tsuyuake | 0.00 | 7.58 ± 1.57 |
| Pi1 | BC3F2 | Tetep | 16.13 | 6.06 ± 1.84 |
| Pi2 | BC3F2 | C101A51 | 64.52 | 3.45 ± 2.35 |
| Pi9 | BC3F2 | 75-1-127 | 96.77 | 1.87 ± 1.18 |
| Pik-R0 | BC2F2 | R03138 | 29.03 | 5.26 ± 1.84 |
| Pik-ME | BC2F2 | Meixiangzhan | 48.34 | 4.29 ± 1.90 |
| Pik-7017 | BC2F2 | IR70175-22-1-1-2-2 | 41.94 | 4.16 ± 1.77 |
Table 4. Disease responses of cloned genes and Pik allele honor plants to Magnaporthe grisea isolates.
| Gene | Generation | Accession a | Resistant rate (%) b | Disease score c |
|---|---|---|---|---|
| CK1 | Parent | Lijiangxintuanheigu | 0.00 | 8.94 ± 0.36 |
| CK2 | Parent | Jin 23B | 3.23 | 7.90 ± 1.54 |
| CK3 | Parent | Kongyu 131 | 0.00 | 7.45 ± 1.84 |
| Pik-m | Parent | Tsuyuake | 77.42 | 3.03 ± 2.59 |
| Pik-s | Parent | Jiahua 1 | 100.00 | 1.71 ± 1.03 |
| Pi1 | Parent | Tetep | 87.10 | 2.55 ± 2.22 |
| Pik-NS | Parent | AMOL3 | 25.81 | 5.90 ± 2.77 |
| Pik-R0 | Parent | R03138 | 96.77 | 0.97 ± 1.14 |
| Pik-ME | Parent | Meixiangzhan | 80.65 | 2.35 ± 2.09 |
| Pik-CE | Parent | IR65482-4-136-2-2-B | 77.42 | 2.48 ± 2.42 |
| Pik-7017 | Parent | IR70175-22-1-1-2-2 | 100.00 | 0.52 ± 0.51 |
| Pik-YD | Parent | Yangdao 4038 | 77.42 | 2.65 ± 2.88 |
| Pik-ZH | Parent | Zhouhui 338 | 96.77 | 4.94 ± 2.10 |
| Pik-m | BC3F2 | Tsuyuake | 0.00 | 7.58 ± 1.57 |
| Pi1 | BC3F2 | Tetep | 16.13 | 6.06 ± 1.84 |
| Pi2 | BC3F2 | C101A51 | 64.52 | 3.45 ± 2.35 |
| Pi9 | BC3F2 | 75-1-127 | 96.77 | 1.87 ± 1.18 |
| Pik-R0 | BC2F2 | R03138 | 29.03 | 5.26 ± 1.84 |
| Pik-ME | BC2F2 | Meixiangzhan | 48.34 | 4.29 ± 1.90 |
| Pik-7017 | BC2F2 | IR70175-22-1-1-2-2 | 41.94 | 4.16 ± 1.77 |
| [1] |
Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J Z, Matsumoto T, Ono K, Yano M. 2008. Two adjacent nucleotide- binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics, 180: 2267-2276.
PMID |
| [2] |
Ashkani S, Rafii M Y, Shabanimofrad M, Miah G, Sahebi M, Azizi P, Tanweer F A, Akhtar M S, Nasehi A. 2015. Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front Plant Sci, 6: 886.
PMID |
| [3] | Chen X W, Shang J J, Chen D X, Lei C L, Zou Y, Zhai W X, Liu G Z, Xu J C, Ling Z Z, Cao G, Ma B T, Wang Y P, Zhao X F, Li S G, Zhu L H. 2006. A β-lectin receptor kinase gene conferring rice blast resistance. Plant J, 46: 794-804. |
| [4] | Chen Z X, Zhao W, Zhu X B, Zou C D, Yin J J, Chern M, Zhou X G, Ying H, Jiang X, Li Y Z, Liao H C, Cheng M P, Li W T, He M, Wang J, Wang J C, Ma B T, Wang J R, Li S G, Zhu L H, Chen X W. 2018. Identification and characterization of rice blast resistance gene Pid4 by a combination of transcriptomic profiling and genome analysis. J Genet Genomics, 45: 663-672. |
| [5] | Dai L Y, Wu J, Li X B, Wang X J, Liu X L, Jantasuriyarat C, Kudrna D, Yu Y, Wing R A, Han B, Zhou B, Wang G L. 2010. Genomic structure and evolution of the Pi2/9 locus in wild rice species. Theor Appl Genet, 121: 295-309. |
| [6] | Deng Y W, Zhai K R, Xie Z, Yang D Y, Zhu X D, Liu J Z, Wang X, Qin P, Yang Y Z, Zhang G M, Li Q, Zhang J F, Wu S Q, Milazzo J, Mao B Z, Wang E T, Xie H A, Tharreau D, He Z H. 2017. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science, 355: 962-965. |
| [7] | Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M. 2009. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science, 325: 998-1001. |
| [8] | Hayashi N, Inoue H, Kato T, Funao T, Shirota M, Shimizu T, Kanamori H, Yamane H, Hayano-Saito Y, Matsumoto T, Yano M, Takatsuji H. 2010. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J, 64: 498-510. |
| [9] | Hittalmani S, Parco A, Mew T V, Zeigler R S, Huang N. 2000. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor Appl Genet, 100: 1121-1128. |
| [10] | Hua L X, Wu J Z, Chen C X, Wu W H, He X Y, Lin F, Wang L, Ashikawa I, Matsumoto T, Wang L, Pan Q H. 2012. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor Appl Genet, 125: 1047-1055. |
| [11] | Inoue H, Nakamura M, Mizubayashi T, Takahashi A, Sugano S, Fukuoka S, Hayashi N. 2017. Panicle blast 1 (Pb1) resistance is dependent on at least four QTLs in the rice genome. Rice, 10: 36. |
| [12] | IRRI. 2002. Standard Evaluation System for Rice. Manila, the Philippine: IRRI. |
| [13] |
Ishihara T, Hayano-Saito Y, Oide S, Ebana K, La N T Hayashi K, Ashizawa T, Suzuki F, Koizumi S. 2014. Quantitative trait locus analysis of resistance to panicle blast in the rice cultivar Miyazakimochi. Rice, 7: 2.
PMID |
| [14] | Jiang J F, Mou T M, Yu H H, Zhou F S. 2015. Molecular breeding of thermo-sensitive genic male sterile (TGMS) lines of rice for blast resistance using Pi2gene. Rice, 8: 11. |
| [15] |
Jiang N, Li Z Q, Wu J, Wang Y, Wu L Q, Wang S H, Wang D, Wen T, Liang Y, Sun P Y, Liu J L, Dai L Y, Wang Z L, Wang C, Luo M Z, Liu X L, Wang G L. 2012. Molecular mapping of the Pi2/9 allelic gene Pi2-2 conferring broad-spectrum resistance to Magnaporthe oryzae in the rice cultivar Jefferson. Rice, 5: 29.
PMID |
| [16] | Lee S K, Song M Y, Seo Y S, Kim H K, Ko S, Cao P J, Suh J P, Yi G, Roh J H, Lee S, An G, Hahn T R, Wang G L, Ronald P, Jeon J S. 2009. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding- leucine-rich repeat genes. Genetics, 181: 1627-1638. |
| [17] | Li W T, Zhu Z W, Chern M, Yin J J, Yang C, Ran L, Cheng M P, He M, Wang K, Wang J, Zhou X G, Zhu X B, Chen Z X, Wang J C, Zhao W, Ma B T, Qin P, Chen W L, Wang Y P, Liu J L, Wang W M, Wu X J, Li P, Wang J R, Zhu L H, Li S G, Chen X W. 2017. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell, 170: 114-126. |
| [18] | Li W T, Chern M, Yin J J, Wang J, Chen X W. 2019. Recent advances in broad-spectrum resistance to the rice blast disease. Curr Opin Plant Biol, 50: 114-120. |
| [19] | Lin F, Chen S, Que Z Q, Wang L, Liu X Q, Pan Q H. 2007. The blast resistance gene Pi37 encodes a nucleotide binding site leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics, 177: 1871-1880. |
| [20] |
Liu G, Lu G, Zeng L, Wang G L. 2002. Two broad-spectrum blast resistance genes, Pi9(t) and Pi2(t), are physically linked on rice chromosome 6. Mol Genet Genomics, 267: 472-480.
PMID |
| [21] | Liu X Q, Lin F, Wang L, Pan Q H. 2007. The in silico map-based cloning of Pi36, a rice coiled-coil nucleotide-binding site leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics, 176: 2541-2549. |
| [22] | Mi J M, Yang D B, Chen Y, Jiang J F, Mou H P, Huang J B, Ouyang Y D, Mou T M. 2018. Accelerated molecular breeding of a novel P/TGMS line with broad-spectrum resistance to rice blast and bacterial blight in two-line hybrid rice. Rice, 11: 11. |
| [23] |
Miah G, Rafii M Y, Ismail M R, Puteh A B, Rahim H A, Asfaliza R, Latif M A. 2013. Blast resistance in rice: A review of conventional breeding to molecular approaches. Mol Biol Rep, 40: 2369-2388.
PMID |
| [24] | Murray M G, Thompson W F. 1980. Rapid isolation of high molecular weight plant. DNA Nucleic Acids Res, 8: 4321-4325. |
| [25] | Qu S H, Liu G F, Zhou B, Bellizzi M, Zeng L R, Dai L Y, Han B, Wang G L. 2006. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics, 172: 1901-1914. |
| [26] |
Sharma T R, Madhav M S, Singh B K, Shanker P, Jana T K, Dalal V, Pandit A, Singh A, Gaikwad K, Upreti H C, Singh N K. 2005. High-resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to Magnaporthe grisea. Mol Genet Genomics, 274: 569-578.
PMID |
| [27] | Shen M, Lin J. 2004. The economic impact of rice blast disease in China. In: Zeigler R S, Leong S A, Teng P S. Rice Blast Disease. Wallingford, UK: CAB International/IRRI: 321-331. |
| [28] |
Skamnioti P, Gurr S J. 2009. Against the grain: Safeguarding rice from rice blast disease. Trends Biotechnol, 27(3): 141-150.
PMID |
| [29] | Tian D G, Chen Z J, Chen Z Q, Zhou Y C, Wang Z H, Wang F, Chen S B. 2016. Allele-specific marker-based assessment revealed that the rice blast resistance genes Pi2 and Pi9 have not been widely deployed in Chinese indica rice cultivars. Rice, 9: 19. |
| [30] | Wang B H, Ebbole D J, Wang Z H. 2017. The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. J Integr Agric, 16: 2746-2760. |
| [31] | Wang G L, Valent B. 2017. Durable resistance to rice blast. Science, 355: 906-907. |
| [32] | Wu K J, Xu T, Guo C J, Zhang X H, Yang S H. 2012. Heterogeneous evolutionary rates of Pi2/9 homologs in rice. BMC Genet, 13: 73. |
| [33] | Wu Y Y, Yu L, Pan C H, Dai Z Y, Li Y H, Xiao N, Zhang X X, Ji H J, Huang N S, Zhao B H, Zhou C H, Liu G Q, Liu X J, Pan X B, Liang C Z, Li A H. 2016. Development of near-isogenic lines with different alleles of Piz locus and analysis of their breeding effect under Yangdao 6 background. Mol Breed, 36(2): 12. |
| [34] | Xiao N, Wu Y Y, Pan C H, Yu L, Chen Y, Liu G Q, Li Y H, Zhang X X, Wang Z P, Dai Z Y, Liang C Z, Li A H. 2017. Improving of rice blast resistances in japonica by pyramiding major R genes. Front Plant Sci, 7: 1918. |
| [35] | Xie Z, Yan B X, Shou J Y, Tang J, Wang X, Zhai K R, Liu J Y, Li Q, Luo M Z, Deng Y W, He Z H. 2019. A nucleotide-binding site-leucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice. Philos Trans R Soc Lond B Biol Sci, 374: 20180308 |
| [36] | Xu X, Lv Q M, Shang J J, Pang Z Q, Zhou Z Z, Wang J, Jiang G H, Tao Y, Xu Q, Li X B, Zhao X F, Li S G, Xu J C, Zhu L H. 2014. Excavation of Pid3orthologs with differential resistance spectra to Magnaporthe oryzae in rice resource. PLoS One, 9(3): e93275. |
| [37] | Yadav M K, Aravindan S, Ngangkham U, Prabhukarthikeyan S R, Keerthana U, Raghu S, Pramesh D, Banerjee A, Roy S, Sanghamitra P, Adak T, Priyadarshinee P, Jena M, Kar M K, Rath P C. 2019. Candidate screening of blast resistance donors for rice breeding. J Genet, 98: 73. |
| [38] | Zhao H J, Wang X Y, Jia Y L, Minkenberg B, Wheatley M, Fan J B, Jia M H, Famoso A, Edwards J D, Wamishe Y, Valent B, Wang G L, Yang Y N. 2018. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat Commun, 9: 2039. |
| [39] | Zhou B, Qu S H, Liu G F, Dolan M, Sakai H, Lu G D, Bellizzi M, Wang G L. 2006. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant-Microbe Interact, 19: 1216-1228. |
| [40] | Zhou X C, Jiang G H, Yang L W, Qiu L, He P, Nong C X, Wang Y Y, He Y Q, Xing Y Z. 2018. Gene diagnosis and targeted breeding for blast-resistant Kongyu 131 without changing regional adaptability. J Genet Genomics, 45: 539-547. |
| [41] | Zhou Y, Lei F, Wang Q, He W C, Yuan B, Yuan W Y. 2020. Identification of novel alleles of the rice blast-resistance gene Pi9 through sequence-based allele mining. Rice, 13: 80. |
| [42] | Zhu Y Y, Chen H R, Fan J H, Wang Y Y, Li Y, Chen J B, Fan J X, Yang S S, Hu L P, Leung H, Mew T W, Teng P S, Wang Z H, Mundt C C. 2000. Genetic diversity and disease control in rice. Nature, 406: 718-722. |
| [1] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [2] | Zhu Jinling, Wei Ruping, Wang Xin, Zheng Chaoqun, Wang Mengmeng, Yang Yicheng, Yang Liuyan. Polyphosphate Accelerates Transformation of Nonstructural Carbohydrates to Improve Growth of ppk-Expressing Transgenic Rice in Phosphorus Deficiency Culture [J]. Rice Science, 2023, 30(3): 235-246. |
| [3] | Wu Zhongling, Qiu Jiehua, Shi Huanbin, Lin Chuyu, Yue Jiangnan, Liu Zhiquan, Xie Wei, Naweed I. Naqvi, Kou Yanjun, Tao Zeng. Polycomb Repressive Complex 2-Mediated H3K27 Trimethylation Is Required for Pathogenicity in Magnaporthe oryzae [J]. Rice Science, 2022, 29(4): 363-374. |
| [4] | Suhas Gorakh Karkute, Vishesh Kumar, Mohd Tasleem, Dwijesh Chandra Mishra, Krishna Kumar Chaturvedi, Anil Rai, Amitha Mithra Sevanthi, Kishor Gaikwad, Tilak Raj Sharma, Amolkumar U. Solanke. Genome-Wide Analysis of von Willebrand Factor A Gene Family in Rice for Its Role in Imparting Biotic Stress Resistance with Emphasis on Rice Blast Disease [J]. Rice Science, 2022, 29(4): 375-384. |
| [5] | Junhua Lu, Xuemei Yang, Jinfeng Chen, Tingting Li, Zijin Hu, Ying Xie, Jinlu Li, Jiqun Zhao, Mei Pu, Hui Feng, Jing Fan, Yanyan Huang, Jiwei Zhang, Wenming Wang, Yan Li. Osa-miR439 Negatively Regulates Rice Immunity Against Magnaporthe oryzae [J]. Rice Science, 2021, 28(2): 156-165. |
| [6] | Yanchang Luo, Tingchen Ma, Teo Joanne, Zhixiang Luo, Zefu Li, Jianbo Yang, Zhongchao Yin. Marker-Assisted Breeding of Thermo-Sensitive Genic Male Sterile Line 1892S for Disease Resistance and Submergence Tolerance [J]. Rice Science, 2021, 28(1): 89-98. |
| [7] | Ning Xiao, Yunyu Wu, Aihong Li. Strategy for Use of Rice Blast Resistance Genes in Rice Molecular Breeding [J]. Rice Science, 2020, 27(4): 263-277. |
| [8] | B. ANGELES-SHIM Rosalyn, P. REYES Vincent, M. del VALLE Marilyn, S. LAPIS Ruby, SHIM Junghyun, SUNOHARA Hidehiko, K. JENA Kshirod, ASHIKARI Motoyuki, DOI Kazuyuki. Marker-Assisted Introgression of Quantitative Resistance Gene pi21 Confers Broad Spectrum Resistance to Rice Blast [J]. Rice Science, 2020, 27(2): 113-123. |
| [9] | Pathaichindachote Wanwarang, Panyawut Natjaree, Sikaewtung Kannika, Patarapuwadol Sujin, Muangprom Amorntip. Genetic Diversity and Allelic Frequency of Selected Thai and Exotic Rice Germplasm Using SSR Markers [J]. Rice Science, 2019, 26(6): 393-403. |
| [10] | Donde Ravindra, Kumar Jitendra, Gouda Gayatri, Kumar Gupta Manoj, Mukherjee Mitadru, Yasin Baksh Sk, Mahadani Pradosh, Kumar Sahoo Khirod, Behera Lambodar, Kumar Dash Sushanta. Assessment of Genetic Diversity of Drought Tolerant and Susceptible Rice Genotypes Using Microsatellite Markers [J]. Rice Science, 2019, 26(4): 239-247. |
| [11] | Kazemi Sheidollah, Reza Eshghizadeh Hamid, Zahedi Morteza. Responses of Four Rice Varieties to Elevated CO2 and Different Salinity Levels [J]. Rice Science, 2018, 25(3): 142-151. |
| [12] | Yaobin Qin, Peng Cheng, Yichen Cheng, Yue Feng, Derun Huang, Tingxu Huang, Xianjun Song, Jiezheng Ying. QTL-Seq Identified a Major QTL for Grain Length and Weight in Rice Using Near Isogenic F2 Population [J]. Rice Science, 2018, 25(3): 121-131. |
| [13] | Xiongsiyee Vua, Rerkasem Benjavan, Veeradittakit Jeeraporn, Saenchai Chorpet, Lordkaew Sittichai, Thebault Prom-u-thai Chanakan. Variation in Grain Quality of Upland Rice from Luang Prabang Province, Lao PDR [J]. Rice Science, 2018, 25(2): 94-102. |
| [14] | Rekha Talukdar Preeti, Rathi Sunayana, Pathak Khanin, Kumar Chetia Sanjay, Nath Sarma Ramendra. Population Structure and Marker-Trait Association in Indigenous Aromatic Rice [J]. Rice Science, 2017, 24(3): 145-154. |
| [15] | Anupam Alpana, Imam Jahangir, Mohammad Quatadah Syed, Siddaiah Anantha, Prasad Das Shankar, Variar Mukund, Prasad Mandal Nimai. Genetic Diversity Analysis of Rice Germplasm in Tripura State of Northeast India Using Drought and Blast Linked Markers [J]. Rice Science, 2017, 24(1): 10-20. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||