
Rice Science ›› 2024, Vol. 31 ›› Issue (6): 659-672.DOI: 10.1016/j.rsci.2024.08.002
• Reviews • Previous Articles Next Articles
Durga Prasad Mullangie1, Kalaimagal Thiyagarajan1( ), Manonmani Swaminathan2, Jagadeesan Ramalingam3, Sritharan Natarajan2, Senthilkumar Govindan2
), Manonmani Swaminathan2, Jagadeesan Ramalingam3, Sritharan Natarajan2, Senthilkumar Govindan2
Received:2024-05-13
Accepted:2024-08-21
Online:2024-11-28
Published:2024-12-10
Contact:
Kalaimagal Thiyagarajan (kalaimagal.t@tnau.ac.in)
Durga Prasad Mullangie, Kalaimagal Thiyagarajan, Manonmani Swaminathan, Jagadeesan Ramalingam, Sritharan Natarajan, Senthilkumar Govindan. Breeding Resilience: Exploring Lodging Resistance Mechanisms in Rice[J]. Rice Science, 2024, 31(6): 659-672.
Add to citation manager EndNote|Ris|BibTeX
| Biochemical | Staining | Wet estimation technique | Reference |
|---|---|---|---|
| Non-reducing sugars | Iodine staining | Anthrone test-glucose | de Bruyn et al, |
| Pectin | Ruthenium red staining; 0.02% unesterified pectin; de-esterified with 0.1 mol/L Na2CO3 overnight at 4o followed by staining | Carbazole method (galacturonic is standard); FTIR; HPLC | Hou et al, |
| Suberin | Sudan red staining | Vapor osmometry; FTIR | Lopes et al, |
| Lignin | Weisner staining (Phloroglucinol staining); Toluidine staining | Karlson lignin; thioglycolic acid; acetyl bromide; DFRC, FTIR, HPLC, AIL, and ASL | Sjöberg et al, |
| Silicon | Digestion with octyl-alcohol, H2O2, and NaOH, and determination by ICP-OES; FTIR | Kraska and Breitenbeck, | |
| Potassium | Digestion with H2O2 and H2SO4 followed by measured through plasma spectrometer | Yue et al, | |
| Cellulose | Calcofluor staining by the use of fluorescent microscope | Updergraff’s method; modified Updergraff’s method; FTIR | Bauer and Ibáñez, |
Table 1. Phenotyping for biochemical estimation.
| Biochemical | Staining | Wet estimation technique | Reference |
|---|---|---|---|
| Non-reducing sugars | Iodine staining | Anthrone test-glucose | de Bruyn et al, |
| Pectin | Ruthenium red staining; 0.02% unesterified pectin; de-esterified with 0.1 mol/L Na2CO3 overnight at 4o followed by staining | Carbazole method (galacturonic is standard); FTIR; HPLC | Hou et al, |
| Suberin | Sudan red staining | Vapor osmometry; FTIR | Lopes et al, |
| Lignin | Weisner staining (Phloroglucinol staining); Toluidine staining | Karlson lignin; thioglycolic acid; acetyl bromide; DFRC, FTIR, HPLC, AIL, and ASL | Sjöberg et al, |
| Silicon | Digestion with octyl-alcohol, H2O2, and NaOH, and determination by ICP-OES; FTIR | Kraska and Breitenbeck, | |
| Potassium | Digestion with H2O2 and H2SO4 followed by measured through plasma spectrometer | Yue et al, | |
| Cellulose | Calcofluor staining by the use of fluorescent microscope | Updergraff’s method; modified Updergraff’s method; FTIR | Bauer and Ibáñez, |
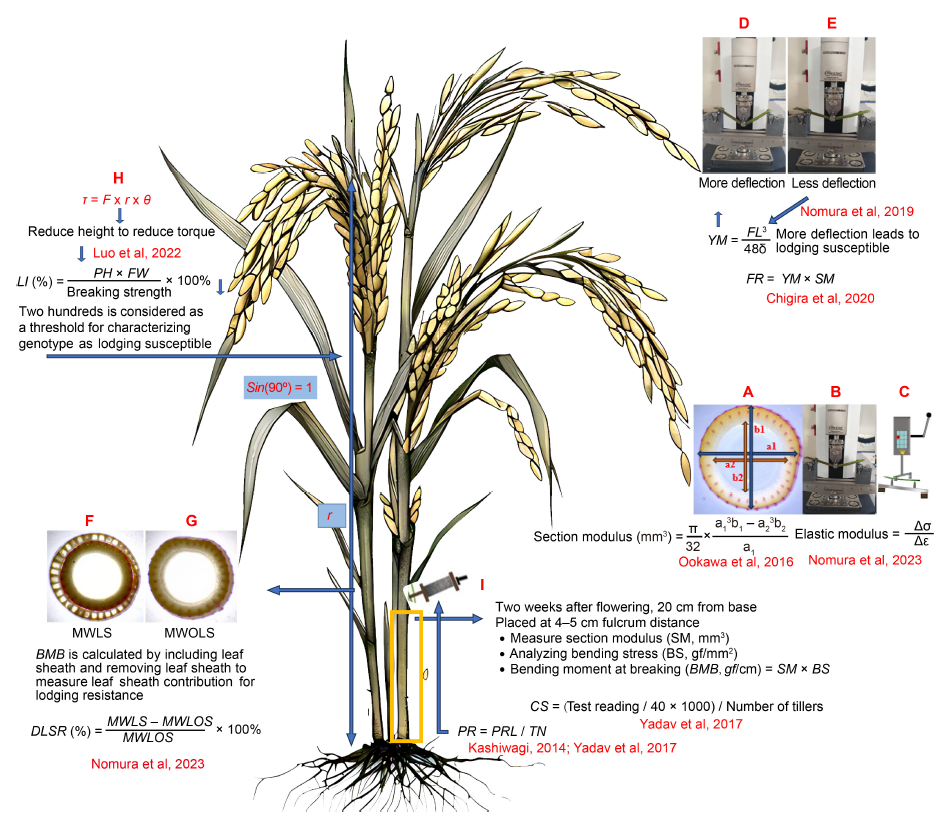
Fig. 1. Mechanics of lodging resistance in rice. A, Section modulus (a1 and b1, the major and minor outer diameters; a2 and b2, the major and minor inner diameters). B, Universal Testing Machine (UTM), used to measure bending stress or load and deflection. C, Plant lodging tester, not as bulky as the UTM used to measure bending stress or load and deflection and elastic modulus. D and E, Deflection of a weak culm is more (D), but a stiff culm is less (E). F and G, Bending moment at breaking (BMB) with leaf sheath (MWLS, F) and without leaf sheath (MWOLS, G) used to measure degree of leaf sheath reinforcement (DLSR) to evaluate the contribution of leaf sheath for lodging resistance. H, Understanding bending-type lodging, less plant height leads to reduced lodging index. I, Prostrate tester utilized to measure bending resistance of the canopy (Pushing resistance reading/tiller number per plant) and culm strength. Δσ, Change in stress; Δε, Change in strain; YM, Young modulus; FL, Load × fulcrum distance; FR, Flexural rigidity; SM, Section modulus; τ, Torque; F, Force; r, Distance from the pivot point; θ, The angle between the force and the lever arm; LI, Lodging index; PH, Plant height; FW, Fresh weight of culm; BS, Bending stress; CS, Culm strength; PR, Pushing resistance; PRL, Pushing resistance of lower internode; TN, Tiller number. The rice image is generated using Meta AI (Llama 3.1).
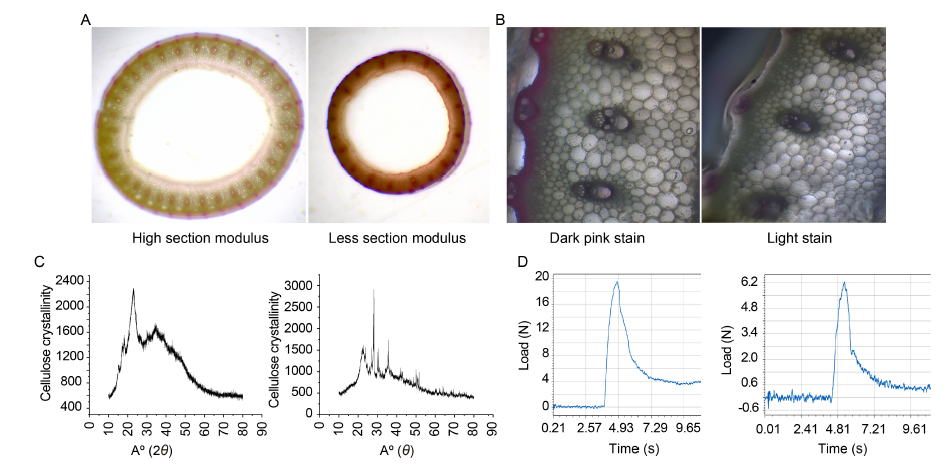
Fig. 2. Comparisons for understanding lodging resistance. A, High section modulus, i.e., less area in the medullary region/lacuna, generally leads to lodging resistance, while low section modulus, i.e., more area in the medullary region, leads to lodging susceptibility. B, Dark pink phloroglucinol stain suggests the presence of more lignin, while light stain signifies the presence of less lignin. C, Powdered X-ray diffraction (XRD) is used to determine cellulose crystallinity. A sharp peak at 22.5º indicates highly crystalline cellulose. Peaks at 22.5º and 18.0º on the X axis are checked for a rough estimation of cellulose crystallinity (sengal method). D, Genotype with high bending stress is observed withstand a load of 18.6 N before breaking, and the genotype with comparatively lower bending stress is observed withstand a load of 6.2 N load before breaking (y-axis, Load; x-axis, Time in seconds).
| Gene | Chr. | Marker | Mapping population | Remark | Reference |
|---|---|---|---|---|---|
| qCD1.1, qCS1.1 | 1 | Id10019973-Id1006772 | Swarna × Moroberekan♂ (BIL) | Lead to an increase in culm diameter and culm strength | Yadav et al, |
| qLR1, qLR8 | 1 | BIN1-161-BIN1-162 BIN8-32-BIN8-33 | 93-11 × O. longistaminata♂ (BIL) | An increase in CD and breaking strength pleiotropic effect qLR1 and qLR8 is seen for lodging resistance | Long et al, |
| OsCYPq1 | 1 | RM12285-RM212 | Cheongcheong♂ × Nagdong (DH) | The candidate gene identified controls basal internode length and mutations in it led to reduced internode elongation and also showed pleiotropic effect by increasing yield | Zhao et al, |
| Gnla | 1 | N321-RM10316 | R498♂ × R3551 (RIL and CSSL) | Increase in CD and crown root development hence can also deal with root lodging | Tu et al, |
| SCM1 | 1 | RM562-RM5 | Habataki♂ × Sasanishiki (CSSL) | Increases culm wall thickness | Yang et al, |
| qSCM4 | 2 | SSR1-RM5511 | LTH♂ × Shennong265 (F2 and RIL) | Increase in CWT, CD, and culm folding resistance | Yang et al, |
| SCM3 | 3 | RM15761-RM15782 SNPV318_03-RM15767 | Koshihikari × Chugoku 117♂ (BIL) | Improves CD, SM, and BMB | Yano et al, |
| prl4 | 4 | RM5749 | Nona Bokra♂ × Koshihikari (CSSL) | Pushing resistance, and accumulates NSC in basal culm | Kashiwagi, |
| prl5 | 5 | C1081 | Nipponbare × Kasalath♂ (NIL) | Delays leaf senescence, PR, NSC content, starch reaccumulation, and high YM | Kashiwagi et al, |
| SCM2 | 6 | RM20546-RM20562 | Sasanishiki × Habataki♂ (CSSL and NIL) | Increases CWT and affects yield because it increases the number of spikelets per panicle | Ookawa et al, |
| qTyM6, qTyH6 | 6 | RM3343-20318 | Cheongcheong♂ × Nagdong (DH) | Resistance reported in Typhoons, as it reduces plant height and lowers plant thrust resistance | Zhao et al, |
| sdm8 | 8 | C10122 | Kasalath♂ × Nipponbare (CSSL) | Increases in CD, CWT, culm stiffness, and heavy lower part of the plant | Kashiwagi et al, |
| BSUC11 | 11 | G44 and G257 | Kasalath♂ × Koshihikari (CSSL) | Increases content of holocellulose in upper culm; thickening of cortical fiber tissue in internode 1 | Kashiwagi et al, |
Table 2. Genes associated with lodging resistance.
| Gene | Chr. | Marker | Mapping population | Remark | Reference |
|---|---|---|---|---|---|
| qCD1.1, qCS1.1 | 1 | Id10019973-Id1006772 | Swarna × Moroberekan♂ (BIL) | Lead to an increase in culm diameter and culm strength | Yadav et al, |
| qLR1, qLR8 | 1 | BIN1-161-BIN1-162 BIN8-32-BIN8-33 | 93-11 × O. longistaminata♂ (BIL) | An increase in CD and breaking strength pleiotropic effect qLR1 and qLR8 is seen for lodging resistance | Long et al, |
| OsCYPq1 | 1 | RM12285-RM212 | Cheongcheong♂ × Nagdong (DH) | The candidate gene identified controls basal internode length and mutations in it led to reduced internode elongation and also showed pleiotropic effect by increasing yield | Zhao et al, |
| Gnla | 1 | N321-RM10316 | R498♂ × R3551 (RIL and CSSL) | Increase in CD and crown root development hence can also deal with root lodging | Tu et al, |
| SCM1 | 1 | RM562-RM5 | Habataki♂ × Sasanishiki (CSSL) | Increases culm wall thickness | Yang et al, |
| qSCM4 | 2 | SSR1-RM5511 | LTH♂ × Shennong265 (F2 and RIL) | Increase in CWT, CD, and culm folding resistance | Yang et al, |
| SCM3 | 3 | RM15761-RM15782 SNPV318_03-RM15767 | Koshihikari × Chugoku 117♂ (BIL) | Improves CD, SM, and BMB | Yano et al, |
| prl4 | 4 | RM5749 | Nona Bokra♂ × Koshihikari (CSSL) | Pushing resistance, and accumulates NSC in basal culm | Kashiwagi, |
| prl5 | 5 | C1081 | Nipponbare × Kasalath♂ (NIL) | Delays leaf senescence, PR, NSC content, starch reaccumulation, and high YM | Kashiwagi et al, |
| SCM2 | 6 | RM20546-RM20562 | Sasanishiki × Habataki♂ (CSSL and NIL) | Increases CWT and affects yield because it increases the number of spikelets per panicle | Ookawa et al, |
| qTyM6, qTyH6 | 6 | RM3343-20318 | Cheongcheong♂ × Nagdong (DH) | Resistance reported in Typhoons, as it reduces plant height and lowers plant thrust resistance | Zhao et al, |
| sdm8 | 8 | C10122 | Kasalath♂ × Nipponbare (CSSL) | Increases in CD, CWT, culm stiffness, and heavy lower part of the plant | Kashiwagi et al, |
| BSUC11 | 11 | G44 and G257 | Kasalath♂ × Koshihikari (CSSL) | Increases content of holocellulose in upper culm; thickening of cortical fiber tissue in internode 1 | Kashiwagi et al, |

Fig. 3. Pleiotropic action of SCM3, SCM2 and depiction of other genes involved in lodging resistance. Gn1a is expressed in culm, roots, inflorescence meristem, and leaves (grey dots). The null allele gn1a in the roots leads to well-developed crown roots, and this allele leads to an increase in grain number. The Gn1a allele SCM1 increases culm diameter and strength. The allele of APO1 (SCM2) increases spikelet number and culm diameter (blue dots). SCM3 decreases tiller number but increases spikelet number (pink dots). The prl4 allele leads to an increase in the bending resistance of the lower internode. GA-inactive (#), GA-active (*), and GA-inactive GA (##) [conversions from active gibberellic acid (GA) to inactive GA alleles SD1 (GA20 oxidase) and SBI (GA2 oxidase)] lead to a reduction in GA levels, leading to dwarfism, prl5 is a gene associated with late senescence, and the orange boxes represent genes involved in lignin and cellulose biosynthesis.The rice image is generated using Meta AI (Llama 3.1).
| [1] | Adjah K L, Asante M D, Toure A, Aziadekey M, Amoako-Andoh F O, Frei M, Diallo Y, Agboka K. 2022. Improvement of rice production under drought conditions in West Africa: Application of QTLs in breeding for drought resistance. Rice Sci, 29(6): 512-521. |
| [2] | Agarie S, Agata W, Uchida H, Kubota F, Kaufman P B. 1996. Function of silica bodies in the epidermal system of rice (Oryza sativa L.): Testing the window hypothesis. J Exp Bot, 47: 655-660. |
| [3] | Alonso-Blanco C, Koornneef M, van Ooijen J W. 2006. QTL analysis. In: Salinas J, Sanchez-Serrano J J. Arabidopsis Protocols. New Jersey, USA: Humana Press: 79-100. |
| [4] | Bauer S, Ibáñez A B. 2014. Rapid determination of cellulose. Biotechnol Bioeng, 111(11): 2355-2357. |
| [5] | Chigira K, Kojima N, Yamasaki M, Yano K, Adachi S, Nomura T, Jiang M J, Katsura K, Ookawa T. 2020. Landraces of temperate japonica rice have superior alleles for improving culm strength associated with lodging resistance. Sci Rep, 10(1): 19855. |
| [6] | Dampanaboina L, Yuan N, Mendu V. 2021a. Estimation of plant biomass lignin content using thioglycolic acid (TGA). J Vis Exp, (173): e62055. |
| [7] | Dampanaboina L, Yuan N, Mendu V. 2021b. Estimation of crystalline cellulose content of plant biomass using the Updegraff method. J Vis Exp, (171): e62031. |
| [8] | de Bruyn J W, van Keulen H A, Ferguson J H A. 1968. Rapid method for the simultaneous determination of glucose and fructose using anthrone reagent. J Sci Food Agric, 19(10): 597-601. |
| [9] | Dhingani R M, Umrania V V, Tomar R S, Parakhia M V, Golakiya B. 2015. Introduction to QTL mapping in plants. Ann Plant Sci, 4(4): 1072-1079. |
| [10] | Dixon R A, Barros J. 2019. Lignin biosynthesis: Old roads revisited and new roads explored. Open Biol, 9(12): 190215. |
| [11] | Dong X L, Zhou Y, Zhang Y Q, Rong F X, Du J H, Hong Z Y, Hu P S, Lü Y S. 2024. OsbZIP01 affects plant growth and development by regulating OsSD1in rice. Rice Sci, 31(1): 77-86. |
| [12] | Dorairaj D, Ismail M R, Sinniah U R, Kar Ban T. 2017. Influence of silicon on growth, yield, and lodging resistance of MR219, a lowland rice of Malaysia. J Plant Nutr, 40(8): 1111-1124. |
| [13] | Duan C R, Wang B C, Wang P Q, Wang D H, Cai S X. 2004. Relationship between the minute structure and the lodging resistance of rice stems. Colloids Surf B: Biointerfaces, 35(3/4): 155-158. |
| [14] | Fan C F, Feng S Q, Huang J F, Wang Y T, Wu L M, Li X K, Wang L Q, Tu Y Y, Xia T, Li J Y, Cai X W, Peng L C. 2017. AtCesA8- driven OsSUS3 expression leads to largely enhanced biomass saccharification and lodging resistance by distinctively altering lignocellulose features in rice. Biotechnol Biofuels, 10: 221. |
| [15] | Fan C F, Li Y, Hu Z, Hu H Z, Wang G Y, Li A, Wang Y M, Tu Y Y, Xia T, Peng L C, Feng S Q. 2018. Ectopic expression of a novel OsExtensin-like gene consistently enhances plant lodging resistance by regulating cell elongation and cell wall thickening in rice. Plant Biotechnol J, 16(1): 254-263. |
| [16] | Fu W Q, Zhao Y J, Zha X R, Ullah J, Ye M, Shah F, Yuan Q H, Wang P, Tao Y, Wu W. 2023. The potential role of zinc and silicon in improving grain yield and lodging resistance of rice (Oryza sativa L.). Agronomy, 14(1): 91. |
| [17] | Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y. 1999. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA, 96(13): 7575-7580. |
| [18] | Gong D K, Zhang X, Yao J P, Dai G J, Yu G X, Zhu Q, Gao Q, Zheng W J. 2021. Synergistic effects of bast fiber seedling film and nano-silicon fertilizer to increase the lodging resistance and yield of rice. Sci Rep, 11(1): 12788. |
| [19] | Grafius J E, Brown H M. 1954. Lodging resistance in Oats1. Agron J, 46(9): 414-418. |
| [20] | Hackett C A, McLean K, Bryan G J. 2013. Linkage analysis and QTL mapping using SNP dosage data in a tetraploid potato mapping population. PLoS One, 8(5): e63939. |
| [21] | Hai L, Guo H J, Xiao S H, Jiang G L, Zhang X Y, Yan C S, Xin Z Y, Jia J Z. 2005. Quantitative trait loci (QTL) of stem strength and related traits in a doubled-haploid population of wheat (Triticum aestivum L.). Euphytica, 141(1): 1-9. |
| [22] | Hall D, Tegström C, Ingvarsson P K. 2010. Using association mapping to dissect the genetic basis of complex traits in plants. Brief Funct Genomics, 9(2): 157-165. |
| [23] | Hirano K, Okuno A, Hobo T, Ordonio R, Shinozaki Y, Asano K, Kitano H, Matsuoka M. 2014. Utilization of stiff culm trait of rice smos1 mutant for increased lodging resistance. PLoS One, 9(7): e96009. |
| [24] | Hirano K, Ordonio R L, Matsuoka M. 2017. Engineering the lodging resistance mechanism of post-Green Revolution rice to meet future demands. Proc Jpn Acad Ser B, 93(4): 220-233. |
| [25] | Hirose T, Ohdan T, Nakamura Y, Terao T. 2006. Expression profiling of genes related to starch synthesis in rice leaf sheaths during the heading period. Physiol Plant, 128(3): 425-435. |
| [26] | Hong W Y, Chen Y J, Huang S H, Li Y Z, Wang Z M, Tang X R, Pan S G, Tian H, Mo Z W. 2022. Optimization of nitrogen- silicon (N-Si) fertilization for grain yield and lodging resistance of early-season indica fragrant rice under different planting methods. Eur J Agron, 136: 126508. |
| [27] | Hou W C, Chang W H, Jiang C M. 1999. Qualitative distinction of carboxyl group distributions in pectins with ruthenium red. Bot Bull Acad Sin, 40: 115-119. |
| [28] | Hu X M, Cui Y T, Dong G J, Feng A H, Wang D Y, Zhao C Y, Zhang Y, Hu J, Zeng D L, Guo L B, Qian Q. 2019. Using CRISPR- Cas9 to generate semi-dwarf rice lines in elite landraces. Sci Rep, 9(1): 19096. |
| [29] | Jung Y J, Kim J H, Lee H J, Kim D H, Yu J, Bae S, Cho Y G, Kang K K. 2020. Generation and transcriptome profiling of Slr1-d7 and Slr1-d8 mutant lines with a new semi-dominant dwarf allele of SLR1 using the CRISPR/Cas9 system in rice. Int J Mol Sci, 21(15): 5492. |
| [30] | Kadirvel P, Senthilvel S, Geethanjali S, Sujatha M, Varaprasad K S. 2015. Genetic markers, trait mapping and marker-assisted selection in plant breeding. In: Bahadur B, Venkat Rajam M, Sahijram L, Krishnamurthy K. Plant Biology and Biotechnology. New Delhi, India: Springer: 65-88. |
| [31] | Kashiwagi T. 2014. Identification of quantitative trait loci for resistance to bending-type lodging in rice (Oryza sativa L.). Euphytica, 198(3): 353-367. |
| [32] | Kashiwagi T, Madoka Y, Hirotsu N, Ishimaru K. 2006. Locus prl5 improves lodging resistance of rice by delaying senescence and increasing carbohydrate reaccumulation. Plant Physiol Biochem, 44(2/3): 152-157. |
| [33] | Kashiwagi T, Togawa E, Hirotsu N, Ishimaru K. 2008. Improvement of lodging resistance with QTLs for stem diameter in rice (Oryza sativa L.). Theor Appl Genet, 117(5): 749-757. |
| [34] | Kashiwagi T, Munakata J, Ishimaru K. 2016. Functional analysis of the lodging resistance QTL BSUC11 on morphological and chemical characteristics in upper culms of rice. Euphytica, 210(2): 233-243. |
| [35] | Kashiwagi T. 2022. Novel QTL for lodging resistance, PRL 4, improves physical properties with high non-structural carbohydrate accumulation of basal clums in rice (Oryza sativa L.). Euphytica, 218(6): 83. |
| [36] | Kraska J E, Breitenbeck G A. 2010. Simple, robust method for quantifying silicon in plant tissue. Commun Soil Sci Plant Anal, 41(17): 2075-2085. |
| [37] | Kyriakidis N B, Psoma E. 2001. Hydrocolloid interferences in the determination of pectin by the carbazole method. J AOAC Int, 84(6): 1947-1949. |
| [38] | Laosutthipong C, Seritrakul P, NaChiangmai P. 2023. Lodging- related gene expression in upland rice varieties from Pala U Village, Thailand. Int J Agric Technol, 19(4): 1557-1590. |
| [39] | Li F C, Xie G S, Huang J F, Zhang R, Li Y, Zhang M M, Wang Y T, Li A, Li X K, Xia T, Qu C C, Hu F, Ragauskas A J, Peng L C. 2017. OsCESA9 conserved-site mutation leads to largely enhanced plant lodging resistance and biomass enzymatic saccharification by reducing cellulose DP and crystallinity in rice. Plant Biotechnol J, 15(9): 1093-1104. |
| [40] | Li Y H, Qian Q, Zhou Y H, Yan M X, Sun L, Zhang M, Fu Z M, Wang Y H, Han B, Pang X M, Chen M S, Li J Y. 2003. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell, 15(9): 2020-2031. |
| [41] | Li Z Z, Deng F, Zhang C, Zhu L, He L H, Zhou T, Lu H, Zhu S L, Zheng Y L, Zhong X Y, Zhou W, Chen Y, Ren W J, Hu J F. 2023. Can ‘relative culm wall thicknesses’ be used to evaluate the lodging resistance of rice? Arch Agron Soil Sci, 69(6): 934-947. |
| [42] | Li F, Zhang M, Guo K, Hu Z, Zhang R, Feng Y, Yi X, Zou W, Wang L, Wu C. 2015. High-level hemicellulose arabinose predominately affects lignocellulose crystallinity for genetically enhancing both plant lodging resistance and biomass enzymatic digestibility in rice mutants. Plant Biotechnol J, 13(4): 514-525. |
| [43] | Liang S J, Li Z Q, Li X J, Xie H G, Zhu R S, Lin J X, Xie H A, Wu H. 2013. Effects of stem structural characters and silicon content on lodging resistance in rice (Oryza sativa L.). Res Crops, 14(3): 621-636. |
| [44] | Liljegren S. 2010. Phloroglucinol stain for lignin. Cold Spring Harb Protoc, 2010(1): pdb.prot4954. |
| [45] | Liu C, Zheng S, Gui J S, Fu C J, Yu H S, Song D L, Shen J H, Qin P, Liu X M, Han B, Yang Y Z, Li L G. 2018. Shortened basal internodes encodes a gibberellin 2-oxidase and contributes to lodging resistance in rice. Mol Plant, 11(2): 288-299. |
| [46] | Liu Q, Yin C S, Li X, He C Q, Ding Z, Du X. 2022. Lodging resistance of rice plants studied from the perspective of culm mechanical properties, carbon framework, free volume, and chemical composition. Sci Rep, 12(1): 20026. |
| [47] | Liu S T, Huang Y W, Xu H, Zhao M H, Xu Q, Li F C. 2018. Genetic enhancement of lodging resistance in rice due to the key cell wall polymer lignin, which affects stem characteristics. Breed Sci, 68(5): 508-515. |
| [48] | Liu Y T, Li T, Jiang Z S, Zeng C H, He R, Qiu J, Lin X L, Peng L M, Song Y P, Zhou D H, Cai Y C, Zhu C L, Fu J R, He H H, Xu J. 2022. Characterization of a novel weak allele of RGA1/D1 and its potential application in rice breeding. Rice Sci, 29(6): 522-534. |
| [49] | Long W X, Dan D, Yuan Z Q, Chen Y P, Jin J, Yang W L, Zhang Z H, Li N W, Li S Q. 2020. Deciphering the genetic basis of lodging resistance in wild rice Oryza longistaminata. Front Plant Sci, 11: 628. |
| [50] | Lopes M H, Gil A M, Silvestre A J, Neto C P. 2000. Composition of suberin extracted upon gradual alkaline methanolysis of Quercus suber L. cork. J Agric Food Chem, 48(2): 383-391. |
| [51] | Luo X Y, Wu Z F, Fu L, Dan Z W, Yuan Z Q, Liang T, Zhu R S, Hu Z L, Wu X T. 2022. Evaluation of lodging resistance in rice based on an optimized parameter from lodging index. Crop Sci, 62(3): 1318-1332. |
| [52] | Lv S W, Lin Z S, Shen J H, Luo L F, Xu Q G, Li L G, Gui J S. 2024. OsTCP19 coordinates inhibition of lignin biosynthesis and promotion of cellulose biosynthesis to modify lodging resistance in rice. J Exp Bot, 75(1): 123-136. |
| [53] | Merugumala G R, Satyanarayana P V, Chamundeswari N, Ravikumar B N V S R, Ramana Rao P V, Pavani L, Deepika V. 2019. Molecular breeding of ‘Swarna’, a mega rice variety for lodging resistance. Mol Breed, 39(4): 55. |
| [54] | Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y. 2002. Positional cloning of rice semidwarfing gene, sd-1: Rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res, 9(1): 11-17. |
| [55] | Nawaz G, Usman B, Zhao N, Han Y, Li Z H, Wang X, Liu Y G, Li R B. 2020. CRISPR/Cas9 directed mutagenesis of OsGA20ox2 in high yielding basmati rice (Oryza sativa L.) line and comparative proteome profiling of unveiled changes triggered by mutations. Int J Mol Sci, 21(17): 6170. |
| [56] | Niu Y N, Chen T X, Zhao C C, Zhou M X. 2021. Improving crop lodging resistance by adjusting plant height and stem strength. Agronomy, 11(12): 2421. |
| [57] | Nomura T, Arakawa N, Yamamoto T, Ueda T, Adachi S, Yonemaru J I, Abe A, Takagi H, Yokoyama T, Ookawa T. 2019. Next generation long-culm rice with superior lodging resistance and high grain yield, Monster Rice 1. PLoS One, 14( 8): e0221424. |
| [58] | Nomura T, Ohkubo S, Nagano A J, Samadi A F, Adachi S, Ookawa T. 2023. Physiological and morphological factors affecting leaf sheath reinforcement and their contribution to lodging resistance in rice. Plant Prod Sci, 26(1): 48-64. |
| [59] | Ookawa T, Hobo T, Yano M, Murata K, Ando T, Miura H, Asano K, Ochiai Y, Ikeda M, Nishitani R, Ebitani T, Ozaki H, Angeles E R, Hirasawa T, Matsuoka M. 2010a. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat Commun, 1: 132. |
| [60] | Ookawa T, Yasuda K, Kato H, Sakai M, Seto M, Sunaga K, Motobayashi T, Tojo S, Hirasawa T. 2010b. Biomass production and lodging resistance in ‘leaf star’, a new long-culm rice forage cultivar. Plant Prod Sci, 13(1): 58-66. |
| [61] | Ookawa T, Inoue K, Matsuoka M, Ebitani T, Takarada T, Yamamoto T, Ueda T, Yokoyama T, Sugiyama C, Nakaba S, Funada R, Kato H, Kanekatsu M, Toyota K, Motobayashi T, Vazirzanjani M, Tojo S, Hirasawa T. 2014. Increased lodging resistance in long-culm, low-lignin gh2 rice for improved feed and bioenergy production. Sci Rep, 4: 6567. |
| [62] | Ookawa T, Aoba R, Yamamoto T, Ueda T, Takai T, Fukuoka S, Ando T, Adachi S, Matsuoka M, Ebitani T, Kato Y, Mulsanti I W, Kishii M, Reynolds M, Piñera F, Kotake T, Kawasaki S, Motobayashi T, Hirasawa T. 2016. Precise estimation of genomic regions controlling lodging resistance using a set of reciprocal chromosome segment substitution lines in rice. Sci Rep, 6: 30572. |
| [63] | Ookawa T, Nomura T, Kamahora E, Jiang M J, Ochiai Y, Samadi A F, Yamaguchi T, Adachi S, Katsura K, Motobayashi T. 2022. Pyramiding of multiple strong-culm genes originating from indica and tropical japonica to the temperate japonica rice. Sci Rep, 12(1): 15400. |
| [64] | Pan J F, Zhao J L, Liu Y Z, Huang N R, Tian K, Shah F, Liang K M, Zhong X H, Liu B. 2019. Optimized nitrogen management enhances lodging resistance of rice and its morpho-anatomical, mechanical, and molecular mechanisms. Sci Rep, 9(1): 20274. |
| [65] | Pradhan Mitra P, Loqué D. 2014. Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J Vis Exp, 87: e51381. |
| [66] | Rao B K R. 2011. Comparison of three digestion methods for total soil potassium estimation in soils of Papua New Guinea derived from varying parent materials. Commun Soil Sci Plant Anal, 42(11): 1259-1265. |
| [67] | Raes J, Rohde A, Christensen J H, van de Peer Y, Boerjan W. 2003. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol, 133(3): 1051-1071. |
| [68] | Ribaut J M, de Vicente M C, Delannay X. 2010. Molecular breeding in developing countries: Challenges and perspectives. Curr Opin Plant Biol, 13(2): 213-218. |
| [69] | Rutger J N. 2009. The induced sd1 mutant and other useful mutant genes in modern rice varieties. In: Shu Q Y. Induced Plant Mutations in the Genomics Era. Proceeding of an International Joint FAO/IAEA Symposium. Vienna, Austria, 2008: 44-47. |
| [70] | Samadi A F, Suzuki H, Ueda T, Yamamoto T, Adachi S, Ookawa T. 2019. Identification of quantitative trait loci for breaking and bending types lodging resistance in rice, using recombinant inbred lines derived from Koshihikari and a strong culm variety, Leaf Star. Plant Growth Regul, 89: 83-98. |
| [71] | Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush G S, Kitano H, Matsuoka M. 2002. A mutant gibberellin-synthesis gene in rice. Nature, 416: 701-702. |
| [72] | Sato Y, Sentoku N, Miura Y, Hirochika H, Kitano H, Matsuoka M. 1999. Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J, 18(4): 992-1002. |
| [73] | Sjöberg G, Nilsson S I, Persson T, Karlsson P. 2004. Degradation of hemicellulose, cellulose and lignin in decomposing spruce needle litter in relation to N. Soil Biol Biochem, 36(11): 1761-1768. |
| [74] | Spielmeyer W, Ellis M H, Chandler P M. 2002. Semidwarf (sd-1), ‘green revolution’ rice, contains a defective gibberellin 20- oxidase gene. Proc Natl Acad Sci USA, 99(13): 9043-9048. |
| [75] | Tu B, Tao Z, Wang S G, Zhou L, Zheng L, Zhang C, Li X Z, Zhang X Y, Yin J J, Zhu X B, Yuan H, Li T, Chen W L, Qin P, Ma B T, Wang Y P, Li S G. 2022. Loss of Gn1a/OsCKX2 confers heavy-panicle rice with excellent lodging resistance. J Integr Plant Biol, 64(1): 23-38. |
| [76] | Wang J, Wang R, Wang Y, Zhang L, Zhang L, Xu Y, Yao S. 2017. Short and Solid Culm/RFL/APO2 for culm development in rice. Plant J, 91(1): 85-96. |
| [77] | Wang Q X, Gao H L, Liu K, Wang H L, Zhang F, Wei L M, Lu K J, Li M M, Shi Y M, Zhao J H, Zhou W, Peng B, Yuan H Y. 2024. CRISPR/Cas9-mediated enhancement of semi-dwarf glutinous traits in elite Xiangdaowan rice (Oryza sativa L.): Targeting SD1 and Wx genes for yield and quality improvement. Front Plant Sci, 15: 1333191. |
| [78] | Wang Y, Wang M H, Yan X, Chen K X, Tian F H, Yang X, Cao L Y, Ruan N, Dang Z J, Yin X L, Huang Y W, Li F C, Xu Q. 2024. The DEP1 mutation improves stem lodging resistance and biomass saccharification by affecting cell wall biosynthesis in rice. Rice, 17(1): 35. |
| [79] | Wang Y H, Xue Y B, Li J Y. 2005. Towards molecular breeding and improvement of rice in China. Trends Plant Sci, 10(12): 610-614. |
| [80] | Waseem M, Ahmad R, Randhawa M A, Aziz T, Mahmood N. 2016. Influence of silicon application on blast incidence and lodging resistance in rice. J Agric Res, 54(3): 435-443. |
| [81] | Wu L M, Zhang W J, Ding Y F, Zhang J W, Cambula E D, Weng F, Liu Z H, Ding C Q, Tang S, Chen L, Wang S H, Li G H. 2017. Shading contributes to the reduction of stem mechanical strength by decreasing cell wall synthesis in japonica rice (Oryza sativa L.). Front Plant Sci, 8: 881. |
| [82] | Wu W B, Wang W, Meadows M E, Yao X F, Peng W. 2019. Cloud-based typhoon-derived paddy rice flooding and lodging detection using multi-temporal Sentinel-1&2. . Front Earth Sci, 13(4): 682-694. |
| [83] | Yadav S, Singh U M, Naik S M, Venkateshwarlu C, Ramayya P J, Raman K A, Sandhu N, Kumar A. 2017. Molecular mapping of QTLs associated with lodging resistance in dry direct-seeded rice (Oryza sativa L.). Front Plant Sci, 8: 1431. |
| [84] | Yang X L, Lai Y C, Wang L Z, Zhao M H, Wang J Y, Li M X, Chi L Y, Lv G Y, Liu Y H, Cui Z B, Li R, Wu L R, Sun B, Zhang X J, Jiang S K. 2023. Isolation of a novel QTL, qSCM4, associated with strong culm affects lodging resistance and panicle branch number in rice. Int J Mol Sci, 24(1): 812. |
| [85] | Yano K, Ookawa T, Aya K, Ochiai Y, Hirasawa T, Ebitani T, Takarada T, Yano M, Yamamoto T, Fukuoka S, Wu J Z, Ando T, Ordonio R L, Hirano K, Matsuoka M. 2015. Isolation of a novel lodging resistance QTL gene involved in strigolactone signaling and its pyramiding with a QTL gene involved in another mechanism. Mol Plant, 8(2): 303-314 |
| [86] | Yue Y, Haiye Y, Xiaokai L, Lei Z, Yuanyuan S. 2023. Prediction of potassium content in rice leaves based on spectral features and random forests. Agronomy, 13(9): 2337. |
| [87] | Zhang W J, Li G H, Yang Y M, Li Q, Zhang J, Liu J Y, Wang S H, Tang S, Ding Y F. 2014. Effects of nitrogen application rate and ratio on lodging resistance of super rice with different genotypes. J Integr Agric, 13(1): 63-72. |
| [88] | Zhang W J, Wu L M, Wu X R, Ding Y F, Li G H, Li J Y, Weng F, Liu Z H, Tang S, Ding C Q, Wang S H. 2016. Lodging resistance of japonica rice (Oryza sativa L.): Morphological and anatomical traits due to top-dressing nitrogen application rates. Rice, 9(1): 31. |
| [89] | Zhao D D, Son J H, Farooq M, Kim K M. 2021. Identification of candidate gene for internode length in rice to enhance resistance to lodging using QTL analysis. Plants, 10(7): 1369. |
| [90] | Zhao D D, Jang Y H, Kim E G, Park J R, Jan R, Lubna, Asaf S, Asif S, Farooq M, Chung H, Kang D J, Kim K M. 2023. Identification of a major locus for lodging resistance to typhoons using QTL analysis in rice. Plants, 12(3): 449. |
| [1] | Fu Yiwei, Wu Jiayelu, Wu Mingming, Ye Shenghai, Zhai Rongrong, Ye Jing, Zhu Guofu, Yu Faming, Lu Yanting, Zhang Xiaoming. Progress on Molecular Mechanism of Heat Tolerance in Rice [J]. Rice Science, 2024, 31(6): 673-687. |
| [2] | Yang Yigang, Xu Ya’nan, Bai Yeran, Zhang Yuanpei, Han Wei, Makoto Saito, Lü Guohua, Song Jiqing, Bai Wenbo. Mixed-Oligosaccharides Promote Seedling Growth of Direct-Seeded Rice under Salt and Alkaline Stress [J]. Rice Science, 2024, 31(6): 712-724. |
| [3] | Ren Jian, Hu Kelin, Feng Puyu, William D. Batchelor, Liu Haitao, Lü Shihua. Simulating Responses of Rice Yield and Nitrogen Fates to Ground Cover Rice Production System under Different Types of Precipitation Years [J]. Rice Science, 2024, 31(6): 725-739. |
| [4] | Tao Yi, Xiao Deshun, Ye Chang, Liu Kancheng, Tang Xinxin, Ma Hengyu, Chu Guang, Yu Kai, Xu Chunmei, Wang Danying. Compound Microbial Agent Improves Soil Redox Status to Reduce Methane Emissions from Paddy Fields [J]. Rice Science, 2024, 31(6): 740-750. |
| [5] | Sitthikorn Bodeerath, Jeeraporn Veeradittakit, Sansanee Jamjod, Chanakan Prom-U-Thai. Applying Boron Fertilizer at Different Growth Stages Promotes Boron Uptake and Productivity in Rice [J]. Rice Science, 2024, 31(6): 751-760. |
| [6] | Kunhikrishnan Hemalatha Dhanyalakshmi, Reshma Mohan, Sasmita Behera, Uday Chand Jha, Debashis Moharana, Ahalya Behera, Sini Thomas, Preman Rejitha Soumya, Rameswar Prasad Sah, Radha Beena. Next Generation Nutrition: Genomic and Molecular Breeding Innovations for Iron and Zinc Biofortification in Rice [J]. Rice Science, 2024, 31(5): 526-544. |
| [7] | Zhang Youliang, Zhu Kaican, Tang Yongqi, Feng Shaoyuan. Rice Cultivation under Film Mulching Can Improve Soil Environment and Be Beneficial for Rice Production in China [J]. Rice Science, 2024, 31(5): 545-555. |
| [8] | Chirag Maheshwari, Nitin Kumar Garg, Archana Singh, Aruna Tyagi. Ameliorative Effects of Paclobutrazol via Physio-Biochemical and Molecular Manifestation in Rice under Water Deficit Stress [J]. Rice Science, 2024, 31(5): 603-616. |
| [9] | Hong Weiyuan, Li Ziqiu, Feng Xiangqian, Qin Jinhua, Wang Aidong, Jin Shichao, Wang Danying, Chen Song. Estimating Key Phenological Dates of Multiple Rice Accessions Using Unmanned Aerial Vehicle-Based Plant Height Dynamics for Breeding [J]. Rice Science, 2024, 31(5): 617-628. |
| [10] | Liyana Sara, Sompop Saeheng, Panupong Puttarak, Lompong Klinnawee. Changes in Metabolites and Allelopathic Effects of Non-Pigmented and Black-Pigmented Lowland Indica Rice Varieties in Phosphorus Deficiency [J]. Rice Science, 2024, 31(4): 434-448. |
| [11] | Zhou Tianshun, Yu Dong, Wu Liubing, Xu Yusheng, Duan Meijuan, Yuan Dingyang. Seed Storability in Rice: Physiological Foundations, Molecular Mechanisms, and Applications in Breeding [J]. Rice Science, 2024, 31(4): 401-416. |
| [12] | Mingyo Ha, Hyo-Young Jeong, Ju Hun Lee, Hyun-Jung Chung. Combined Insights from Leachate Structure and Microstructure Characteristics for Eating Quality of Convenience Rice Processed by Super-Heated and Pressurized Steam Technologies [J]. Rice Science, 2024, 31(4): 475-488. |
| [13] | Anuradha Kumari, Wusirika Ramakrishna. Anticancer Activity of Rice Callus Suspension Cultures from Aromatic Varieties and Metabolites Regulated in Treated Cancer Cell Lines [J]. Rice Science, 2024, 31(4): 449-462. |
| [14] | Priyanka Negi, Jagadish Rane, Rajendra Sadashiv Wagh, Tukaram Jayaram Bhor, Dipti Digambar Godse, Priyanka Jadhav, C. Anilkumar, Dasari Sreekanth, K. Sammi Reddy, Sharad Ramrao Gadakh, K. M. Boraih, C. B. Harisha, P. S. Basavaraj. Direct-Seeded Rice: Genetic Improvement of Game-Changing Traits for Better Adaption [J]. Rice Science, 2024, 31(4): 417-433. |
| [15] | Zhang Fengmin, Cao Zhenzhen, Zheng Xin, He Yuntao, Chen Mingxue, Lin Xiaoyan. Interaction Between Ustilaginoidea virens and Rice and Its Sustainable Control [J]. Rice Science, 2024, 31(3): 269-284. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||