
Rice Science ›› 2019, Vol. 26 ›› Issue (2): 77-87.DOI: 10.1016/j.rsci.2018.07.001
• Research Papers • Previous Articles Next Articles
Shufen Chao1,,#, Yicong Cai1,,#, Baobing Feng1, Guiai Jiao1, Zhonghua Sheng1, Ju Luo1, Shaoqing Tang1,2, Jianlong Wang2, Peisong Hu1( ), Xiangjin Wei1(
), Xiangjin Wei1( )
)
Received:2018-06-01
Accepted:2018-07-27
Online:2019-03-04
Published:2018-12-18
Shufen Chao, Yicong Cai, Baobing Feng, Guiai Jiao, Zhonghua Sheng, Ju Luo, Shaoqing Tang, Jianlong Wang, Peisong Hu, Xiangjin Wei. Editing of Rice Isoamylase Gene ISA1 Provides Insights into Its Function in Starch Formation[J]. Rice Science, 2019, 26(2): 77-87.
Add to citation manager EndNote|Ris|BibTeX
| Primer name | Primer sequence |
|---|---|
| Target1-F | CAGTGGACGGCGTGAGCACGATC |
| Target1-R | AACGATCGTGCTCACGCCGTCCA |
| Sq-primer | CTCCTTCCTTCCGTCCACTTCATC |
| Hyg-F | GCTGTTATGCGGCCATTGTC |
| Hyg-R | GACGTCTGTCGAGAAGTTTC |
| Cas9-F | GAGACAAGCCGTTTCGTGG |
| Cas9-R | ATCAATCCGTTCTTGCCAG |
| Sq-primer | CTCCTTCCTTCCGTCCACTTCATC |
| Actin-F | TGCTATGTACGTCGCCATCCA |
| Actin-R | AATGAGTAACCACGCTCCGTC |
Supplemental Table 1 Primers used in this study
| Primer name | Primer sequence |
|---|---|
| Target1-F | CAGTGGACGGCGTGAGCACGATC |
| Target1-R | AACGATCGTGCTCACGCCGTCCA |
| Sq-primer | CTCCTTCCTTCCGTCCACTTCATC |
| Hyg-F | GCTGTTATGCGGCCATTGTC |
| Hyg-R | GACGTCTGTCGAGAAGTTTC |
| Cas9-F | GAGACAAGCCGTTTCGTGG |
| Cas9-R | ATCAATCCGTTCTTGCCAG |
| Sq-primer | CTCCTTCCTTCCGTCCACTTCATC |
| Actin-F | TGCTATGTACGTCGCCATCCA |
| Actin-R | AATGAGTAACCACGCTCCGTC |
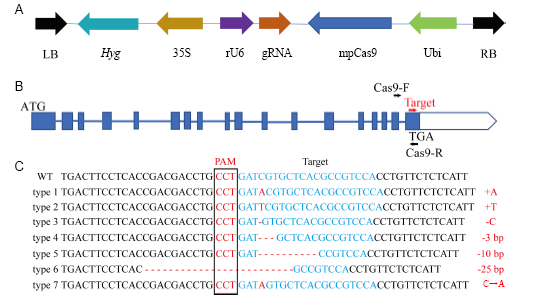
Fig. 1. CRISPR/Cas9 mediated editing of ISA1.A, The structure of the T-DNA region of the Cas9/guide RNV (gRNA) vector. Marker gene Hygromycin (Hyg) was driven by the CaMV35S (35S) promoter whereas the gRNA was driven by the rice U6 promoter and the mpCas9 was driven by the Ubiquitin (Ubi) promoter. LB, Left border; RB, Right border. B, The structure of ISA1 gene. The primer pairs (Cas9-F and Cas9-R) were used to amplify the region that was sequenced in the genotypes. C, Identification of generated mutation forms in ISA1 by sequencing of the target site (protospacer adjacent motif region) in T0 transgenic events. PAM, Protospacer adjacent motif. The mutation forms in all 36 T0 transgenic events can be divided into 7 categories. Homozygous mutations included type 1 to type 6, while a heterozygous mutation was classified as type 7. Type 1 and type 3 mutations were selected for further analysis.

Supplemental Fig. 1. Amino acid sequence alignment of all mutation forms.Different background colors indicate different identities of multiple sequence alignments by DNAMAN. Residues in white with blue background means 100% identity; and those in black color with red background indicates 80% identity and those in black color with emerald blue background indicate 60% identity.
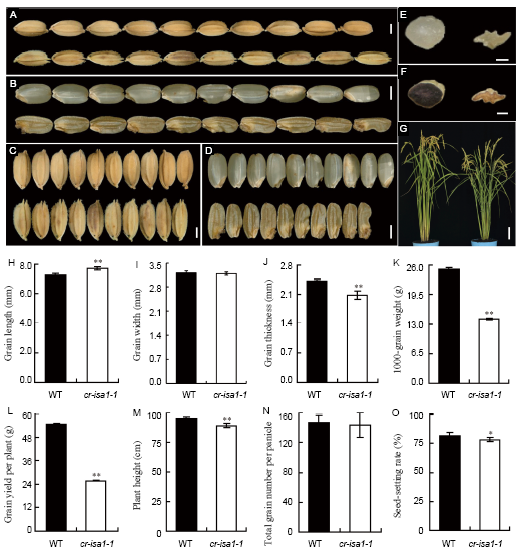
Fig. 2. Phenotypes of the wild-type (WT) and cr-isa1-1 mutant (T1 generation).A, Appearance of mature seeds for WT (above) and cr-isa1-1 (below). B, Appearance of brown rice for WT (above) and cr-isa1-1 (below). C, Grain widths of WT (above) and cr-isa1-1 (below). D, Brown rice widths of WT (above) and the cr-isa1-1 transgenic line (below). E, Cross sections of WT (left) and cr-isa1-1 transgenic line (right) brown rice. F, Iodine staining of brown rice in WT (left) and cr-isa1-1 transgenic line (right). G, Representative plants of WT (left) and cr-isa1-1 (right) after heading. H, Comparison of grain length. I, Comparison of grain width. J, Comparison of grain thickness. K, Comparison of 1000-grain weight. L, Comparison of grain yield per plant. M, Comparison of plant height. N, Comparison of total grain number per panicle. O, Comparison of seed-setting rate. Scale bars are 2 mm in A to D, 1 mm in E and F, and 10 cm in G. Values in H to O are Mean ± SD from three biological replicates, with no less than 50, 50, 50 and 200 seeds in each replication for H, I, J and K, and no less than 5 plants for L to O, respectively. Asterisks indicate statistical significance by the Student’s t-test (*, P < 0.05; **, P < 0.01).
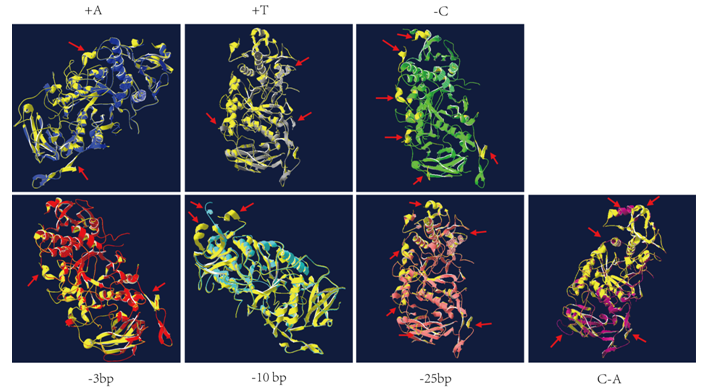
Supplemental Fig. 2 The predicted three-dimensional protein structure of all mutation forms which was compared by using the tool Swiss-PdbViewer 4.1.0. The structure in yellow color represented the wild-type and structures in other colors all stand for the lines of cr-isa1. The red arrows indicated the differences of protein structure between wild-type and each cr-isa1 lines.
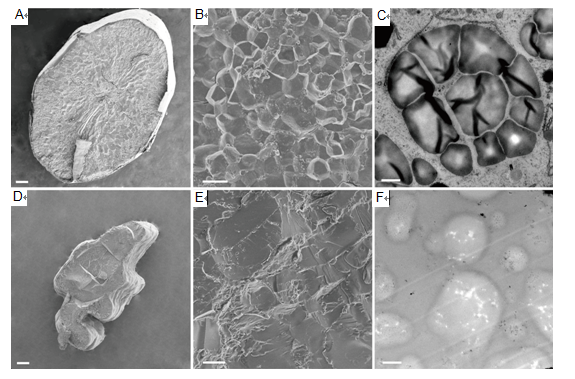
Fig. 3. Electron microscopy images of wild-type and cr-isa1-1 (T1 generation).A, Scanning electron microscopy (SEM) analysis of mature endosperm of wild-type. B, Central region of mature endosperm in the wild-type. C, Amyloplast in endosperm cells of wild-type at 10 d after flowering visualized by transmission electron microscopy analysis. D, SEM analysis of mature endosperm of the cr-isa1-1 transgenic plant. E, Central region of mature endosperm in cr-isa1-1. F, Amyloplast in endosperm cells of cr-isa1-1 at 10 d after flowering visualized by transmission electron microscopy analysis. Scale bars are 0.2 mm in A and D, 10 μm in B and E, and 1 μm in C and F.
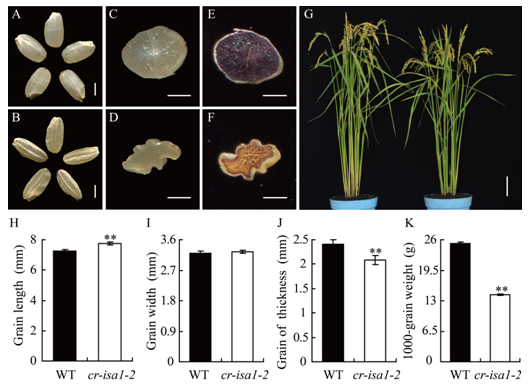
Supplemental Fig. 3 Phenotype of the cr-isa1-2 mutant (T1 generation).A, Appearance of the wild-type (WT). B, Appearance of cr-isa1-2. C, Cross section of WT brown rice. D, Cross section of cr-isa1-2 brown rice. E, Iodine stained phenotype of brown rice of WT. F, Iodine stained phenotype of brown rice of cr-isa1-2. G, Representative plants of WT and cr-isa1-2 after heading. H, Grain length, I, Grain width, J, Grain thickness. K, 1000-grain weight. Scale bar: 2 mm in A-B, 1 mm in C-F and 10 cm in G. Values are Mean ± SD with three biological replicates, at least 50, 10 and 200 seeds in each replication in H, I, J and K, respectively. Asterisks in H-K indicate statistical significance compared with the WT, as determined by Student’s t-test (*P < 0.05, **P < 0.01).
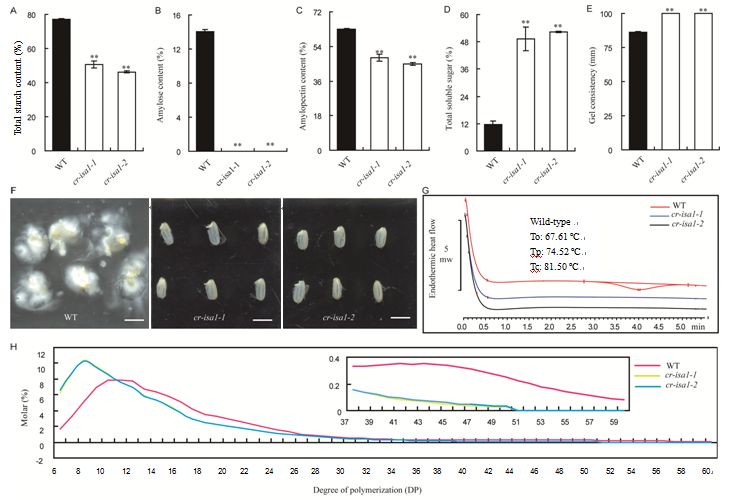
Fig. 4. Starch physicochemical characteristics analysis in T1 generation of cr-isa1 mutants.A, Total starch content. B, Amylose content. C, Amylopectin content. D, Total soluble sugar. E, Gel consistency. F, Starch solubility in KOH solution (Scale bar is 5 mm). G, Thermal characteristics presented by modulated differential scanning calorimetry (MDSC) curves. To, Onset temperature, Tp, Peak temperature; Tc, Conclusion temperature. H, Chain length distributions of amylopectin in wild-type (WT) and cr-isa1. The picture in the upper right corner shows the enlarged region with degree of polymerization ranging from 37 to 60. Data are shown as Mean ± SD (n = 3). Asterisks indicate the statistical significance by the Student’s t-test at the 0.01 level.
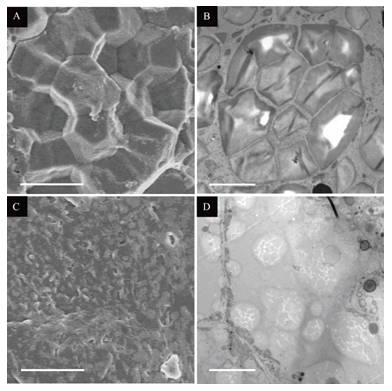
Supplemental Fig. 4 Electron microscopy images of cr-isa1-2 (T1) endosperm.A, SEM image of the central region of mature endosperm of the wild-type. B, TEM image of an amyloplast in 10 DAF endosperm cells of the wild-type. C, SEM image of the central region of mature endosperm of cr-isa1-2. D, TEM image of an amyloplast in 10 DAF endosperm cells of cr-isa1-2. Scale bar: 5?m. SEM, Scanning electron microscopy; DAF, days after fertilization; TEM, Transmission electron microscopy.
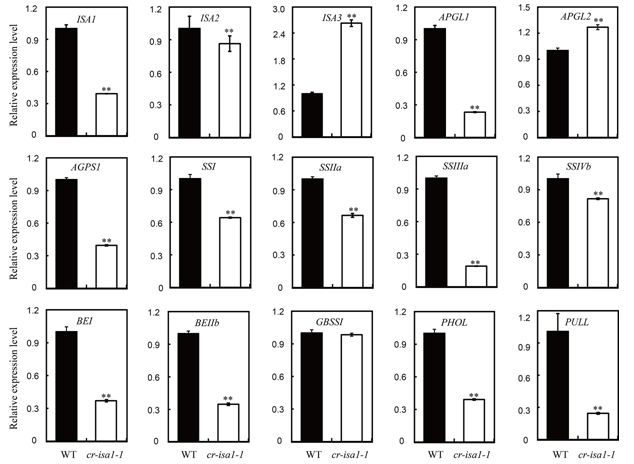
Fig. 5. Relative expression levels of starch metabolism related genes in seeds at 10 d after flowering in wild-type (WT) and cr-isa1-1 (T1).Total RNA extracted from developing seeds at 10 d after flowering was used for quantitative real-time PCR analysis. Expression level of each gene in the wild-type was set as reference value of 1. Data are Mean ± SD (n = 3). Asterisks indicate the statistical significance between wild-type and the cr-isa1-1 as determined by the Student’s t-test at the 0.01 level.
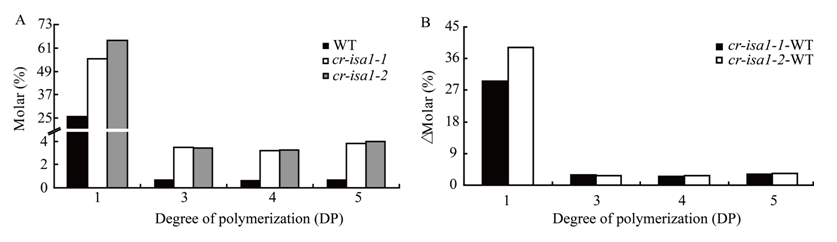
Supplemental Fig. 5 Short chain length distributions of amylopectin in cr-isa1.A, Chain length distributions in the range of DP 1-5 of amylopectin in WT and cr-isa1 mutants. B, Differences in the amylopectin chain length distributions in the range of DP 1-5 between cr-isa1 and the WT.
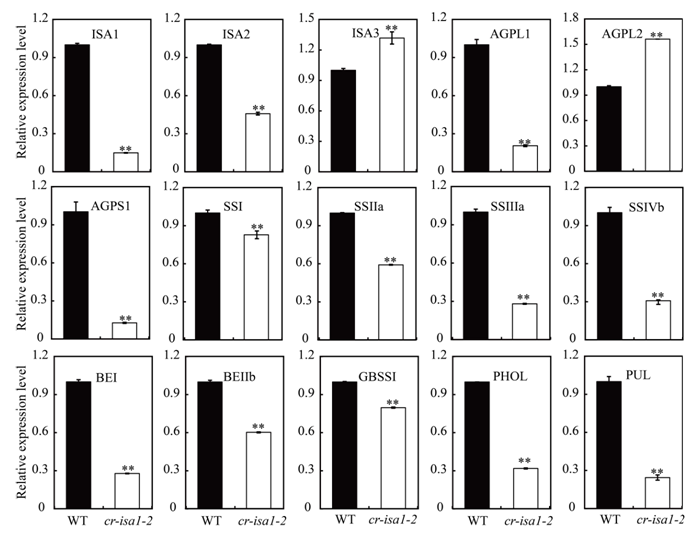
Supplemental Fig. 6 Expression level of starch metabolism related genes in wild-type and cr-isa1-2 (T1) 10 DAF seeds.Total RNA was extracted from 10 DAF developing seeds for qRT-PCR analysis. Expression level of each gene in the wild-type was set as reference value of 1. Data are displayed as means ± SD with three biological replicates. Asterisks indicate the statistical significance between wild-type and the cr-isa1-2 as determined by a Student’s t-test (*P < 0.05; **P < 0.01). DAF, Days after fertilization.
| [1] | Akdogan G, Kubota J, Kubo A, Takaha T, Kitamura S.2011. Expression and characterization of rice disproportionating enzymes.J Appl Glycosci, 58(3): 99-105. |
| [2] | Burton R A, Jenner H, Carrangis L, Fahy B, Fincher G B, Hylton C, Laurie D A, Parker M, Waite D, van Wegen S, Verhoeven T, Denyer K.2002. Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity.Plant J, 31(1): 97-112. |
| [3] | Delatte T, Trevisan M, Parker M L, Zeeman S C.2005. Arabidopsis mutantsAtisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J, 41(6): 815-830. |
| [4] | Erlander S R.1958. A proposed mechanism for the synthesis of starch from glycogen.Enzymologia, 19(5): 273-283. |
| [5] | Ferreira S J, Senning M, Fischer-Stettler M, Streb S, Ast M, Neuhaus H E, Zeeman S C, Sonnewald S, Sonnewald U.2017. Simultaneous silencing of isoamylases ISA1, ISA2 and ISA3 by multi-target RNAi in potato tubers leads to decreased starch content and an early sprouting phenotype. PLoS One, 12(7): e0181444. |
| [6] | Fitzgerald M A, Mccouch S R, Hall R D.2009. Not just a grain of rice: The quest for quality.Trends Plant Sci, 14(3): 133-139. |
| [7] | Fujita N, Kubo A, Suh D S, Wong K S, Jane J L, Ozawa K, Takaiwa F, Inaba Y, Nakamura Y.2003. Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm.Plant Cell Physiol, 44(6): 607-618. |
| [8] | Fujita N.2015. Manipulation of rice starch properties for application. In: Nakamura Y. Starch Metabolism and Structure. Tokyo: Springer: 342-343. |
| [9] | Han Y P, Xu M L, Liu X Y, Yan C J, Korban S S, Chen X L, Gu M H.2004. Genes coding for starch branching enzymes are major contributors to starch viscosity characteristics in waxy rice (Oryza sativa L.). Plant Sci, 166(2): 357-364. |
| [10] | Haugen T H, Ishaque A, Preiss J.1976. Biosynthesis of bacterial glycogen: Characterization of the subunit structure of Escherichia coli B glucose-1-phosphate adenylyltransferase (EC 2.7.7.27). J Biol Chem, 251(24): 7880-7885. |
| [11] | Hiei Y, Komari T, Kubo T.1997. Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol Biol, 35: 205-218. |
| [12] | Hwang S K, Koper K, Satoh H, Okita T W.2016. Rice endosperm starch phosphorylase (Pho1) assembles with disproportionating enzyme (Dpe1) to form a protein complex that enhances synthesis of malto-oligosaccharides.J Biol Chem, 291: 19994-20007. |
| [13] | James M G, Robertson D S, Myers A M.1995. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell, 7(4): 417-429. |
| [14] | Jeon J S, Ryoo N, Hahn T R, Walla H, Nakamura Y.2010. Starch biosynthesis in cereal endosperm.Plant Physiol Biochem, 48(6): 383-392. |
| [15] | Jones H D.2015. Regulatory uncertainty over genome editing. Nat Plants, 1(1): 14011. |
| [16] | Kang H G, Park S, Matsuoka M, An G.2005. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J, 42(6): 901-911. |
| [17] | Kawagoe Y, Kubo A, Satoh H, Takaiwa F, Nakamura Y.2005. Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm.Plant J, 42(2): 164-174. |
| [18] | Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y.1999. The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm.Plant Physiol, 121(2): 399-410. |
| [19] | Kubo A, Rahman S, Utsumi Y, Li Z, Mukai Y, Yamamoto M, Ugaki M, Harada K, Satoh H, Konik-Rose C, Morell M, Nakamura Y.2005. Complementation of sugary-1 phenotype in rice endosperm with the wheat isoamylase1 gene supports a direct role for isoamylase1 in amylopectin biosynthesis. Plant Physiol, 137(1): 43-56. |
| [20] | Kweon M, Haynes L, Slade L, Levine H.2000. The effect of heat and moisture treatments on enzyme digestibility of AeWx, Aewx and aeWx corn starch.J Therm Anal Cal, 59: 571-586. |
| [21] | Lee S K, Hwang S K, Han M, Eom J S, Kang H G, Han Y, Choi S B, Cho M H, Bhoo S H, An G, Hahn T R, Okita T W, Jeon J S.2007. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol Biol, 65(4): 531-546. |
| [22] | Li J, Huang L, Yao Y, Ma Y Q.2010. The progress in the highly branched carbohydrate polymer phytoglycogen.Acad Period Farm Prod Proc, 4: 35-38. (in Chinese with English abstract) |
| [23] | Li S F, Wei X J, Ren Y L, Qiu J H, Jiao G A, Guo X P, Tang S Q, Wan J M, Hu P S.2017. OsBT1 encodes an ADP-glucose transporter involved in starch synthesis and compound granule formation in rice endosperm.Sci Rep, 7: 40124. |
| [24] | Liu L L, Ma X D, Liu S J, Zhu C L, Jiang L, Wang Y H, Shen Y, Ren Y L, Dong H, Chen L M, Liu X, Zhao Z G, Zhai H Q, Wan J M.2009. Identification and characterization of a novel waxy allele from a Yunnan rice landrace.Plant Mol Biol, 71(6): 609-626. |
| [25] | Liu W Z, Xie X R, Ma X L, Li J, Chen J H, Liu Y G.2015. DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations.Mol Plant, 8(9): 1431-1433. |
| [26] | Ma X L, Chen L T, Zhu Q L, Chen Y L, Liu Y G.2015. Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products.Mol Plant, 8(8): 1285-1287. |
| [27] | Masuko T, Minami A, Iwasaki N, Majita T, Nishimura S I, Lee Y C.2005. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem, 339(1): 69-72. |
| [28] | Mizuno K, Kimura K, Arai Y, Kawasaki T, Shimada H, Baba T.1992. Starch branching enzymes from immature rice seeds.J Biochem, 112(5): 643-651. |
| [29] | Naito Y, Hino K, Bono H, Ui-Tei K.2015. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites.Bioinformatics, 31(7): 1120-1123. |
| [30] | Nakamura Y, Takeichi T, Kawaguchi K, Yamanouchi H.1992. Purification of two forms of starch branching enzyme (Q-enzyme) from developing rice endosperm.Physiol Plant, 84(3): 329-335. |
| [31] | Nakamura Y, Umemoto T, Ogata N, Kuboki Y, Yano M, Sasaki T.1996. Starch debranching enzyme (R-enzyme or pullulanase) from developing rice endosperm: Purification, cDNA and chromosomal localisation of the gene.Planta, 199(2): 209-218. |
| [32] | Peng C, Wang Y H, Liu F, Ren Y L, Zhou K N, Lv J, Zheng M, Zhao S L, Zhang L, Wang C M, Jiang L, Zhang X, Guo X P, Bao Y Q, Wan J M.2014. FLOURY ENDOSPERM 6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm.Plant J, 77(6): 917-930. |
| [33] | Pfister B, Zeeman S C.2016. Formation of starch in plant cell.Cell Mol Life Sci, 73(14): 2781-2807. |
| [34] | Rao Y C, Li Y Y, Qian Q.2014. Recent progress on molecular breeding of rice in China.Plant Cell Rep, 33(4): 551-564. |
| [35] | Ryoo N, Yu C, Park C S, Baik M Y, Park I M, Cho M H, Bhoo S H, Ah G, Hahn T R, Jeon J S.2007. Knockout of a starch synthase geneOsSSIIIa/Flo5 causes white-core floury endosperm in rice(Oryza sativa L.). Plant Cell Rep, 26(7): 1083-1095. |
| [36] | Schmittgen T D, Livak K J.2008. Analyzing real-time PCR data by the comparative CT method.Nat Protoc, 3(6): 1101-1108. |
| [37] | Sestili F, Sparla F, Botticella E, Janni M, DʼOvidio R, Falini G, Marri L, Cuesta-Seijo J A, Moscatello S, Battistelli A, Trost P, Lafiandra D.2016. The down-regulation of the genes encoding isoamylase 1 alters the starch composition of the durum wheat grain.Plant Sci, 252: 230-238. |
| [38] | Shao G N, Xie L H, Jiao G A, Wei X J, Sheng Z H, Tang S Q, Hu P S.2017. CRISPR/CAS9-mediated editing of the fragrant gene Badh2 in rice. Chin J Rice Sci, 31(2): 216-222. (in Chinese with English abstract) |
| [39] | She K C, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M, Tsuge T, Matsumoto K, Kudoh M, Itoh E, Kikuchi S, Kishimoto N, Yazaki J, Ando T, Yano M, Aoyama T, Sasaki T, Satoh H, Shimada H.2010. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell, 22(10): 3280-3294. |
| [40] | Smith A M, Denyer K, Martin C.1997. The synthesis of the starch granule.Annu Rev Plant Physiol Plant Mol Biol, 48: 67-87. |
| [41] | Streb S, Delatte T, Umhang M, Eicke S, Schorderet M, Reinhardt D, Zeeman S C.2008. Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell, 20(12): 3448-3466. |
| [42] | Sun T, Tong L G, Zhao S Y, Wang H W, Han Y F, Zhang Z C, Jin Z X.2018. Effects of nitrogen fertilizer application on starch quality, activities and gene expression levels of related enzymes in rice endosperm.Chin J Rice Sci, 32(5): 475-484. (in Chinese with English abstract) |
| [43] | Sun Y W, Jiao G A, Liu Z P, Zhang X, Li J Y, Guo X P, Du W M, Du J L, Francis F, Zhao Y D, Xia L Q.2017. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front Plant Sci, 8: 298. |
| [44] | Thitisaksakul M, Jimenez R C, Arias M C, Beckles D M.2012. Effects of environmental factors on cereal starch biosynthesis and composition.J Cereal Sci, 56(1): 67-80. |
| [45] | Thompson D B.2000. On the non-random nature of amylopectin branching.Carbohyd Polym, 43(3): 223-239. |
| [46] | Utsumi Y, Nakamura Y.2006. Structural and enzymatic characterization of the isoamylase1 homo-oligomer and the Isoamylase1-isoamylase2 hetro-oligomer from rice endosperm.Planta, 225(1): 75-87. |
| [47] | Utsumi Y, Utsumi C, Sawada T, Fujita N, Nakamura Y.2011. Functional diversity of isoamylase oligomers: The ISA1 homo- oligomer is essential for amylopectin biosynthesis in rice endosperm.Plant Physiol, 156(1): 61-77. |
| [48] | Vrinten P L, Nakamura T.2000. Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues. Plant Physiol, 122(1): 255-264. |
| [49] | Wattebled F, Dong Y, Dumez S, Delvallé D, Planchot V, Berbezy P, Vyas D, Colonna P, Chatterjee M, Ball S, DʼHulst C.2005. Mutants ofArabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol, 138(1): 184-195. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIU Tingting, ZOU Jinpeng, YANG Xi, WANG Kejian, RAO Yuchun, WANG Chun. Development and Application of Prime Editing in Plants [J]. Rice Science, 2023, 30(6): 3-. |
| [7] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [8] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [9] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [10] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [11] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [12] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [13] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [14] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [15] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||