
Rice Science ›› 2019, Vol. 26 ›› Issue (2): 98-108.DOI: 10.1016/j.rsci.2018.12.005
• Research Papers • Previous Articles Next Articles
Bo Wang1,2,#, Zhaohui Zhong2,#, Huanhuan Zhang1, Xia Wang1, Binglin Liu2, Lijia Yang2, Xiangyan Han1, Deshui Yu1, Xuelian Zheng2, Chunguo Wang1, Wenqin Song1, Chengbin Chen1( ), Yong Zhang2(
), Yong Zhang2( )
)
Received:2018-10-23
Accepted:2018-12-27
Online:2019-03-04
Published:2018-12-18
Bo Wang, Zhaohui Zhong, Huanhuan Zhang, Xia Wang, Binglin Liu, Lijia Yang, Xiangyan Han, Deshui Yu, Xuelian Zheng, Chunguo Wang, Wenqin Song, Chengbin Chen, Yong Zhang. Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice[J]. Rice Science, 2019, 26(2): 98-108.
Add to citation manager EndNote|Ris|BibTeX
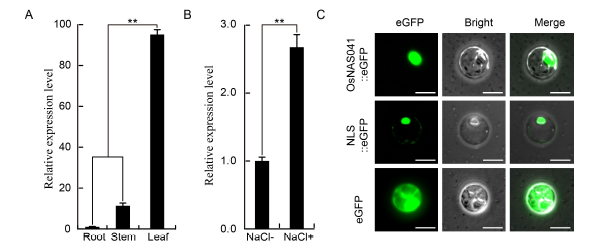
Fig. 1. Expression profile and subcellular location analysis of OsNAC041.A, Quantitative real-time PCR (qRT-PCR) analysis of OsNAC041 expression in samples of root, stem and leaf from two-week-old wild-type (WT) plants. B, qRT-PCR analysis of OsNAC041 expression patterns in WT plants before (NaCl-) and after (NaCl+) salt stress. C, Nuclear localization of OsNAC041 protein in the rice protoplast. NLS, Nuclear localization signal. Scale bar = 20 µm. Data represent Mean ± SE (n = 3). **, Significance at the 0.01 level.
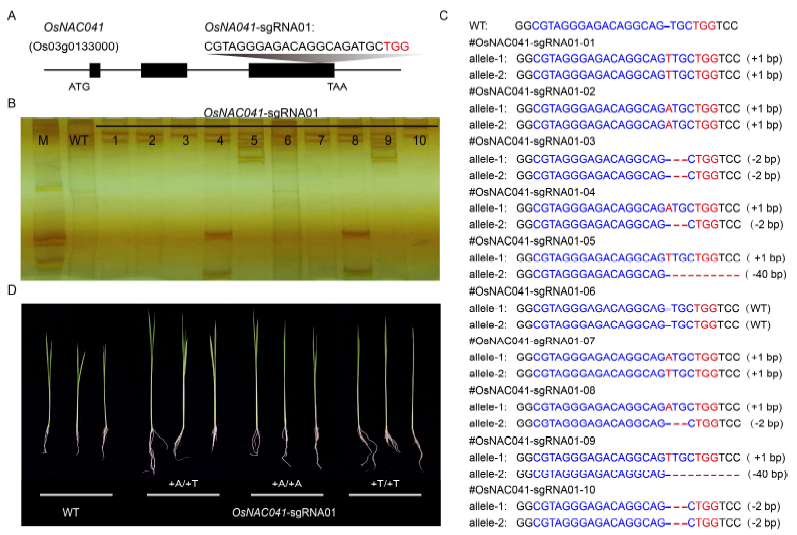
Fig. 2. Targeted mutagenesis of the OsNAC041 locus using CRISPR-Cas9 system.A, Design of sgRNA sites for the OsNAC041 exons. B, Single-strand conformation polymorphism analysis of 10 independent OsNAC041- sgRNA01 T0 lines. M, Marker; WT, Wild-type. C, Sanger sequencing of the target site in the OsNAC041-sgRNA01 T0 lines. D, Phenotypic analysis of OsNAC041 T1 mutant lines under normal condition.
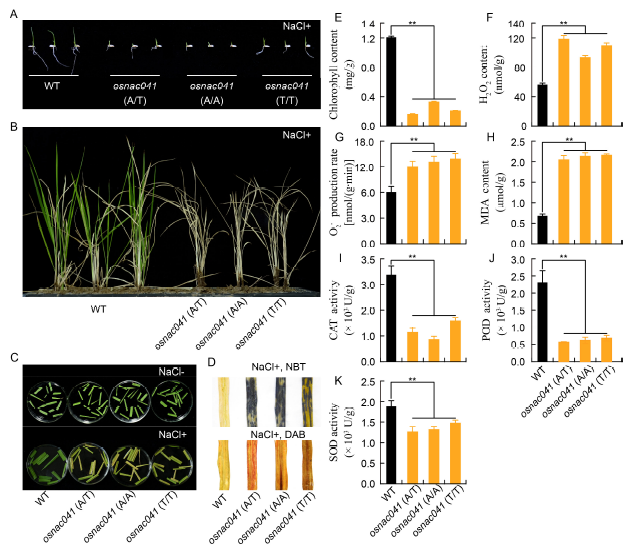
Fig. 3. OsNAC041 targeted mutants showed the salt-sensitive phenotype.A, Germination of OsNAC041 T1 mutant and wild-type (WT) seeds grown on 1/2 MS medium supplemented with NaCl (150 mmol/L). B, Phenotypic analysis of OsNAC041 T1 mutant lines under salt stress. C, Growth of OsNAC041 T1 mutant lines in plugs. D, Levels of O2- and H2O2 in WT and OsNAC041 T1 mutant lines subjected to salt stress. Salt-stressed leaf samples were incubated in nitro-blue tetrazolium (NBT) or diaminobenzidine (DAB) solution. E, Chlorophyll content after 15-day salt stress. F, H2O2 content after 15-day salt stress. G, O2- production rate after 15-day salt stress. H, Malondialdehyde (MDA) content after 15-day salt stress. I, Catalase (CAT) activity after 15-day salt stress. J, Peroxidase (POD) activity after 15-day salt stress. K, Superoxide dismutase (SOD) activity after 15-day salt stress. Data represent Mean ± SE (n = 3). **, Significant at the 0.01 level.
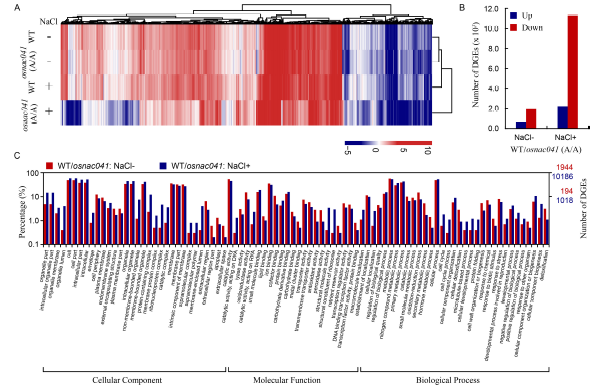
Fig. 4. Targeted knockout of OsNAC041 resulted in global gene expression changes in rice.A, Number of differentially expressed genes (DEGs) in the wild-type (WT) and osnac041 (A/A) T1 mutant lines, based on expression profiles obtained by RNA-Seq. B, Clustering analysis of DEGs in WT and osnac041 T1 mutant lines. Targeted knockout of osnac041 resulted in global changes compared with the WT both with and without salt stress. The color scale corresponds to log2(FPKM) values of the genes. C, Gene ontology classification of DEGs in the following two comparisons: WT and osnac041 T1 mutant lines under normal and salt stress conditions. x-axis shows user selected GO terms; y-axis shows the percentages of genes (number of a particular gene divided by total gene number).
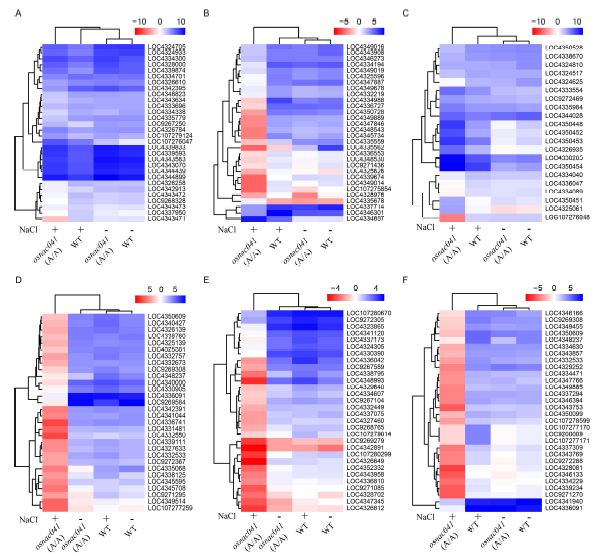
Fig. 5. Transcriptome analysis of genes systemically regulated in the wild-type (WT) and osnac041 T1 mutant lines in response to salt stress.A, Photosynthesis-related genes. B, Peroxisome-related genes. C, Water deprivation-related genes. D, Mitogen-activated protein kinase signal-related genes. E, Eukaryotic-type ABC transporter-related genes. F, Plant hormone regulatory pathway-related genes. Log2(Fold change, FC) of differentially expressed genes in WT and osnac041 T1 mutant lines before (NaCl-) and after (NaCl+) salt stress is defined as [-1.5 > log(FC) > 1.5] with false discovery rate (FDR) < 0.05, and the top 30 most significant differences are shown in the heat-map.
| Predicted bipartite NLS position | Sequence | Score |
| 17 | DLPGFRFHPTEEELLDFYLSRVVLGKKLHFNI | 2.4 |
| 37 | RVVLGKKLHFNIIGTLNIYRHDPWDLPGMAKI | 2.4 |
| 37 | RVVLGKKLHFNIIGTLNIYRHDPWDLPGMAKIGE | 2.1 |
| 91 | RTTERGFWKATGSDRAIRSSGDPKRVIGLKKTLV | 2.2 |
| 91 | RTTERGFWKATGSDRAIRSSGDPKRVIGLKKTLVFY | 2.9 |
| 115 | RVIGLKKTLVFYQGRAPRGTKTDWVMNEYRLPD | 2 |
| 141 | NEYRLPDYGAARAAAPPPKEDMVLCKIYR | 2.9 |
| 161 | DMVLCKIYRKATPLKELEQRASAMEEMQRGS | 2.4 |
| NLS, Nuclear localization signal. | ||
Supplemental Table 1 Result of the NLS-mapper predictions of the location of OsNAC041 expression.
| Predicted bipartite NLS position | Sequence | Score |
| 17 | DLPGFRFHPTEEELLDFYLSRVVLGKKLHFNI | 2.4 |
| 37 | RVVLGKKLHFNIIGTLNIYRHDPWDLPGMAKI | 2.4 |
| 37 | RVVLGKKLHFNIIGTLNIYRHDPWDLPGMAKIGE | 2.1 |
| 91 | RTTERGFWKATGSDRAIRSSGDPKRVIGLKKTLV | 2.2 |
| 91 | RTTERGFWKATGSDRAIRSSGDPKRVIGLKKTLVFY | 2.9 |
| 115 | RVIGLKKTLVFYQGRAPRGTKTDWVMNEYRLPD | 2 |
| 141 | NEYRLPDYGAARAAAPPPKEDMVLCKIYR | 2.9 |
| 161 | DMVLCKIYRKATPLKELEQRASAMEEMQRGS | 2.4 |
| NLS, Nuclear localization signal. | ||
| Name | Primer sequence |
| OsActin-RT-F | AGCTGCGGGTATCCATGAGA |
| OsActin-RT-R | GCAATGCCAGGGAACATAGTG |
| 4324705-RT-F | GTTCTTCGTCCAGGCCATCGTCA |
| 4324705-RT-R | GTAGGCCCATGCGTTGTTGTTGA |
| 4324706-RT-F | GTCTTCGTCAGTGGCAACCTC |
| 4324706-RT-R | GAAACATCTGGCTGAACCTGAG |
| 4324707-RT-F | GGTCGCGTAGTTCAAGTGGG |
| 4324707-RT-R | TCGCCTGACATTATTTCACCAA |
| 4324708-RT-F | TGGACTCGCTGGACCAGAAC |
| 4324708-RT-R | CGGTGTAGGGCTGGAACG |
| 4324709-RT-F | GCAGACGCAGACCAAGAAGAA |
| 4324709-RT-R | GGCGGCAGAGGAAAGAGC |
| 4324710-RT-F | GCCACCATCGAGAAGACGAAGCG |
| 4324710-RT-R | AGCAGGCCGAACTTGCCGTAG |
| 4324711-RT-F | GTGGTTCGGTTCGCCAAGGTG |
| 4324711-RT-R | ATCTCGTCCAGGGCGTAGTTGT |
| 4324712-RT-F | TTGAAGGGATAGGAAGGACC |
| 4324712-RT-R | AGACAGAGTCAGAAGGGAAG |
| 4324713-RT-F | ACTGGAGGAATGTGGGACTGGC |
| 4324713-RT-R | TCACCGACCCGAACAGCAAA |
| 4324714-RT-F | CGCAGCTCCGACAACATCT |
| 4324714-RT-R | GCTGGGTGACACTCTCTCTACAAG |
| OsNAC041-sgRNA01-F | GTGTGCGTAGGGAGACAGGCAGTGC |
| OsNAC041-sgRNA01-R | AAACGCACTGCCTGTCTCCCTACGC |
| NAC041-F | GCAAAATGATCGGAATGAAGTG |
| NAC041-R | GCGAAGACCAAAAGATTGATTG |
Supplemental Table 2 Oligonucleotides used in this study
| Name | Primer sequence |
| OsActin-RT-F | AGCTGCGGGTATCCATGAGA |
| OsActin-RT-R | GCAATGCCAGGGAACATAGTG |
| 4324705-RT-F | GTTCTTCGTCCAGGCCATCGTCA |
| 4324705-RT-R | GTAGGCCCATGCGTTGTTGTTGA |
| 4324706-RT-F | GTCTTCGTCAGTGGCAACCTC |
| 4324706-RT-R | GAAACATCTGGCTGAACCTGAG |
| 4324707-RT-F | GGTCGCGTAGTTCAAGTGGG |
| 4324707-RT-R | TCGCCTGACATTATTTCACCAA |
| 4324708-RT-F | TGGACTCGCTGGACCAGAAC |
| 4324708-RT-R | CGGTGTAGGGCTGGAACG |
| 4324709-RT-F | GCAGACGCAGACCAAGAAGAA |
| 4324709-RT-R | GGCGGCAGAGGAAAGAGC |
| 4324710-RT-F | GCCACCATCGAGAAGACGAAGCG |
| 4324710-RT-R | AGCAGGCCGAACTTGCCGTAG |
| 4324711-RT-F | GTGGTTCGGTTCGCCAAGGTG |
| 4324711-RT-R | ATCTCGTCCAGGGCGTAGTTGT |
| 4324712-RT-F | TTGAAGGGATAGGAAGGACC |
| 4324712-RT-R | AGACAGAGTCAGAAGGGAAG |
| 4324713-RT-F | ACTGGAGGAATGTGGGACTGGC |
| 4324713-RT-R | TCACCGACCCGAACAGCAAA |
| 4324714-RT-F | CGCAGCTCCGACAACATCT |
| 4324714-RT-R | GCTGGGTGACACTCTCTCTACAAG |
| OsNAC041-sgRNA01-F | GTGTGCGTAGGGAGACAGGCAGTGC |
| OsNAC041-sgRNA01-R | AAACGCACTGCCTGTCTCCCTACGC |
| NAC041-F | GCAAAATGATCGGAATGAAGTG |
| NAC041-R | GCGAAGACCAAAAGATTGATTG |
| [1] | Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M.1997. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell, 9(6): 841-857. |
| [2] | Archer E K.2014. American society of plant biologists: Position statement on the education of young children about plants.Cbe-Life Sci Educ, 13(4): 575-576. |
| [3] | Cabello J V, Lodeyro A F, Zurbriggen M D.2014. Novel perspectives for the engineering of abiotic stress tolerance in plants.Curr Opin Biotech, 26: 62-70. |
| [4] | Chen X, Wang Y F, Lv B, Li J, Luo L Q, Lu S C, Zhang X, Ma H, Ming F.2014. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol, 55(3): 604-619. |
| [5] | Depuydt S, Hardtke C S.2011. Hormone signalling crosstalk in plant growth regulation.Curr Biol, 21(9): 365-373. |
| [6] | Doczi R, Okresz L, Romero A E, Paccanaro A, Bogre L.2012. Exploring the evolutionary path of plant MAPK networks.Trends Plant Sci, 17(9): 518-525. |
| [7] | Dolferus R.2014. To grow or not to grow: A stressful decision for plants.Plant Sci, 229: 247-261. |
| [8] | Duan M, Zhang R X, Zhu F G, Zhang Z Q, Gou L M, Wen J Q, Dong J L, Wang T.2017. A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress.Plant Cell, 29(7): 1748-1772. |
| [9] | Ernst H A, Olsen A N, Larsen S, lo Leggio L.2004. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors.EMBO Rep, 5(3): 297-303. |
| [10] | Fang Y J, Liao K F, Du H, Xu Y, Song H Z, Li X H, Xiong L Z.2015. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J Exp Bot, 66: 6803-6817. |
| [11] | Francisco R M, Regalado A, Ageorges A, Burla B J, Bassin B, Eisenach C, Zarrouk O, Vialet S, Marlin T, Chaves M M, Martinoia E, Nagya R.2013. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell, 25(5): 1840-1854. |
| [12] | Hayashi H, Sakamoto A, Alia, Murata N.1998. Enhancement of stress tolerance by gene-engineering of betaine accumulation in plants.Photosynth: Mech Eff: 2419-2424. |
| [13] | Hu H H, Dai M Q, Yao J L, Xiao B Z, Li X H, Zhang Q F, Xiong L Z.2006. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice.Proc Natl Acad Sci USA, 103: 12987-12992. |
| [14] | Hu H H, You J, Fang Y J, Zhu X Y, Qi Z Y, Xiong L Z.2008. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol, 67: 169-181. |
| [15] | Hu H H, Xiong L Z.2014. Genetic engineering and breeding of drought-resistant crops.Annu Rev Plant Biol, 65(1): 715-741. |
| [16] | Jeong J S, Kim Y S, Redillas M C F R, Jang G, Jung H, Bang S W, Choi Y D, Ha S H, Reuzeau C, Kim J K.2013. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field.Plant Biotechnol J, 11(1): 101-114. |
| [17] | Kato H, Motomura T, Komeda Y, Saito T, Kato A.2010. Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J Plant Physiol, 167(7): 571-577. |
| [18] | Kosugi S, Hasebe M, Tomita M, Yanagawa H.2009. Systematic identification of yeast cell cycle-dependent nucleocytoplasmic shuttling proteins by prediction of composite motifs.Proc Natl Acad Sci USA, 106: 10171-10176. |
| [19] | Kou L L, Hu H C, Ma L, Ke X N, Liu M Y, Lian W M, Jin K, Xie L J, Liu Q P.2018. Functional analysis of a copper/zinc SOD encoding gene in response to arsenite stress in rice.Chin J Rice Sci, 32(5): 437-444. (in Chinese with English abstract) |
| [20] | Kudo M, Kidokoro S, Yoshida T, Mizoi J, Todaka D, Fernie A R, Shinozaki K, Yamaguchi-Shinozaki K.2017. Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants.Plant Biotechnol J, 15(4): 458-471. |
| [21] | Kunieda T, Mitsuda N, Ohme-Takagi M, Takeda S, Aida M, Tasaka M, Kondo M, Nishimura M, Hara-Nishimura I.2008. NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell, 20(10): 2631-2642. |
| [22] | Lata C, Prasad M.2011. Role of DREBs in regulation of abiotic stress responses in plants.J Exp Bot, 62(14): 4731-4748. |
| [23] | Lee D K, Chung P J, Jeong J S, Jang G, Bang S W, Jung H, Kim Y S, Ha S H, Choi Y D, Kim J K.2017. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol J, 15(6): 754-764. |
| [24] | Lefevre F, Baijot A, Boutry M.2015. Plant ABC transporters: Time for biochemistry?Biochem Soc T, 43(5): 931-936. |
| [25] | Liang C Z, Wang Y Q, Zhu Y N, Tang J Y, Hu B, Liu L C, Ou S J, Wu H K, Sun X H, Chu J F, Chu C C.2014. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice.Proc Natl Acad Sci USA, 111: 10013-10018. |
| [26] | Liu X M, Nguyen X C, Kim K E, Han H J, Yoo J, Lee K, Kim M C, Yun D J, Chung W S.2013. Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem Biophys Res Comm, 430(3): 1054-1059. |
| [27] | Ma C Q, Wang Y G, Gu D, Nan J D, Chen S X, Li H Y.2017. Overexpression of S-adenosyl-L-methionine synthetase 2 from sugar beet M14 increased Arabidopsis tolerance to salt and oxidative stress. Int J Mol Sci, 18(4): 847. |
| [28] | Mentewab A, Stewart C N.2005. Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat Biotechnol, 23(9): 1177-1180. |
| [29] | Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R.2010. Reactive oxygen species homeostasis and signalling during drought and salinity stresses.Plant Cell Environ, 33(4): 453-467. |
| [30] | Munoz-Munoz J L, Garcia-Molina F, Garcia-Ruiz P A, Arribas E, Tudela J, Garcia-Canovas F, Rodriguez-Lopez J N.2009. Enzymatic and chemical oxidation of trihydroxylated phenols.Food Chem, 113(2): 435-444. |
| [31] | Nuruzzaman M, Sharoni A M, Satoh K, Moumeni A, Venuprasad R, Serraj R, Kumar A, Leung H, Attia K, Kikuchi S.2012. Comprehensive gene expression analysis of the NAC gene family under normal growth conditions, hormone treatment, and drought stress conditions in rice using near-isogenic lines (NILs) generated from crossing Aday selection (drought tolerant) and IR64.Mol Genet Genomics, 287(5): 389-410. |
| [32] | Peleg Z, Blumwald E.2011. Hormone balance and abiotic stress tolerance in crop plants.Curr Opin Plant Biol, 14(3): 290-295. |
| [33] | Petricka J J, Winter C M, Benfey P N.2012. Control of Arabidopsis root development. Annu Rev Plant Biol, 63: 563-590. |
| [34] | Pitzschke A, Datta S, Persak H.2014. Salt stress in Arabidopsis: Lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Mol Plant, 7(4): 722-738. |
| [35] | Rai G K, Rai N P, Rathaur S, Kumar S, Singh M.2013. Expression of rd29A::AtDREB1A/CBF3 in tomato alleviates drought induced oxidative stress by regulating key enzymatic and non- enzymatic antioxidants. Plant Physiol Biochem, 69: 90-100. |
| [36] | Redillas M C F R, Jeong J S, Kim Y S, Jung H, Bang S W, Choi Y D, Ha S H, Reuzeau C, Kim J K.2012. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J, 10(7): 792-805. |
| [37] | Schmidt R, Mieulet D, Hubberten H M, Obata T, Hoefgen R, Fernie A R, Fisahn J, Segundo B S, Guiderdoni E, Schippers J H M, Mueller-Roeber B.2013. SALT-RESPONSIVE ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice.Plant Cell, 25(6): 2115-2131. |
| [38] | Shen H S, Liu C T, Zhang Y, Meng X P, Zhou X, Chu C C, Wang X P.2012. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice.Plant Mol Biol, 80(3): 241-253. |
| [39] | Shen J B, Lv B, Luo L Q, He J M, Mao C J, Xi D D, Ming F.2017. The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci Rep, 7: 40641. |
| [40] | Shi Y T, Ding Y L, Yang S H.2015. Cold signal transduction and its interplay with phytohormones during cold acclimation.Plant Cell Physiol, 56(1): 7-15. |
| [41] | Souer E, van Houwelingen A, Kloos D, Mol J, Koes R.1996. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries.Cell, 85(2): 159-170. |
| [42] | Sousa R H V, Carvalho F E L, Ribeiro C W, Passaia G, Cunha J R, Lima-Melo Y, Margis-Pinheiro M, Silveira J A G.2015. Peroxisomal APX knockdown triggers antioxidant mechanisms favourable for coping with high photorespiratory H2O2 induced by CAT deficiency in rice.Plant Cell Environ, 38: 499-513. |
| [43] | Tang X, Zheng X L, Qi Y P, Zhang D W, Cheng Y, Tang A T, Voytas D F, Zhang Y.2016. A single transcript CRISPR-Cas9 system for efficient genome editing in plants.Mol Plant, 9(7): 1088-1091. |
| [44] | Tang X, Lowder L G, Zhang T, Malzahn A A, Zheng X L, Voytas D F, Zhong Z H, Chen Y Y, Ren Q R, Li Q, Kirkland E R, Zhang Y, Qi Y P.2017. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants.Nat Plants, 3: 17108. |
| [45] | Theodoulou F L, Kerr I D.2015. ABC transporter research: Going strong 40 years on.Biochem Soc Trans, 43(5): 1033-1040. |
| [46] | Tian X J, Li X F, Zhou W J, Ren Y K, Wang Z Y, Liu Z Q, Tang J Q, Tong H N, Fang J, Bu Q Y.2017. Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiol, 175(3): 1337-1349. |
| [47] | Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S.2006. Early infection of scutellum tissue with Agrobacterium allows high- speed transformation of rice. Plant J, 47: 969-976. |
| [48] | Wu Y C, Liu C L, Kuang J, Ge Q, Zhang Y, Wang Z Z.2014. Overexpression of SmLEA enhances salt and drought tolerance in Escherichia coli and Salvia miltiorrhiza. Protoplasma, 251: 1191-1199. |
| [49] | Xiong H Y, Yu J P, Miao J L, Li J J, Zhang H L, Wang X, Liu P L, Zhao Y, Jiang C H, Yin Z G, Li Y, Guo Y, Fu B Y, Wang W S, Li Z K, Ali J, Li Z C.2018. Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiol, 178(1): 451-467. |
| [50] | Xu Z Y, Kim S Y, Hyeon D Y, Kim D H, Dong T, Park Y, Jin J B, Joo S H, Kim S K, Hong J C, Hwang D, Hwang I.2013. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell, 25(11): 4708-4724. |
| [51] | Zheng X N, Chen B, Lu G J, Han B.2009. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance.Biochem Biophy Res Co, 379(4): 985-989. |
| [52] | Zheng X L, Yang S X, Zhang D W, Zhong Z H, Tang X, Deng K J, Zhou J P, Qi Y P, Zhang Y.2016. Effective screen of CRISPR/ Cas9-induced mutants in rice by single-strand conformation polymorphism.Plant Cell Rep, 35(7): 1545-1554. |
| [53] | Zhong Z H, Zhang Y X, You Q, Tang X, Ren Q R, Liu S S, Yang L J, Wang Y, Liu X P, Liu B L, Zhang T, Zheng X L, Le Y, Zhang Y, Qi Y P.2018. Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered PAM sites.Mol Plant, 11(7): 999-1002. |
| [54] | Zhou J P, Deng K J, Cheng Y, Zhong Z H, Tian L, Tang X, Tang A T, Zheng X L, Zhang T, Qi Y P, Zhang Y.2017. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice.Front Plant Sci, 8: 1598. |
| [55] | Zhou M Q, Xu M, Wu L H, Shen C, Ma H, Lin J.2014. CbCBF from Capsella bursa-pastoris enhances cold tolerance and restrains growth in Nicotiana tabacum by antagonizing with gibberellin and affecting cell cycle signaling. Plant Mol Biol, 85(3): 259-275. |
| [56] | Zhu J K.2002. Salt and drought stress signal transduction in plants.Annu Rev Plant Biol, 53: 247-273. |
| [1] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [2] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [3] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [4] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [5] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [6] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [7] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [8] | LIU Tingting, ZOU Jinpeng, YANG Xi, WANG Kejian, RAO Yuchun, WANG Chun. Development and Application of Prime Editing in Plants [J]. Rice Science, 2023, 30(6): 3-. |
| [9] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [10] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [11] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [12] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [13] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||