
Rice Science ›› 2021, Vol. 28 ›› Issue (1): 69-80.DOI: 10.1016/j.rsci.2020.11.008
• Research Papers • Previous Articles Next Articles
Mao-Sen Liu, Shih-Hsuan Tseng, Ting-Chu Chen, Mei-Chu Chung( )
)
Received:2019-10-07
Accepted:2020-03-20
Online:2021-01-28
Published:2021-01-28
Mao-Sen Liu, Shih-Hsuan Tseng, Ting-Chu Chen, Mei-Chu Chung. Visualizing Meiotic Chromosome Pairing and Segregation in Interspecific Hybrids of Rice by Genomic in situ Hybridization[J]. Rice Science, 2021, 28(1): 69-80.
Add to citation manager EndNote|Ris|BibTeX
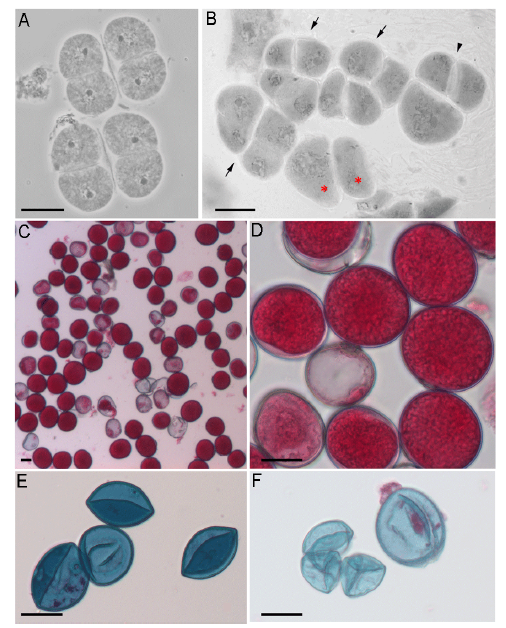
Fig. 1. Viability of pollen grains from interspecific hybrids.A, Tetrad of hybrid IR64 × O. longistaminata (Ol).B, Meiotic end-products of the hybrid O. australiensis (Oa) × O. ridleyi (Or) including tetrad (arrow), triad (arrowhead) and dyad (*). C-F, Pollen viability test by Alexander staining. Viable pollens were solid and stained magenta-red, and non-viable pollens with empty and shriveled features were stained blue. C and D, Pollens of the hybrid IR64 × Ol; E, Pollens of the hybrid Oa × Or; F, Pollens of the hybrid Om × Oa.Scale bars, 20 µm.
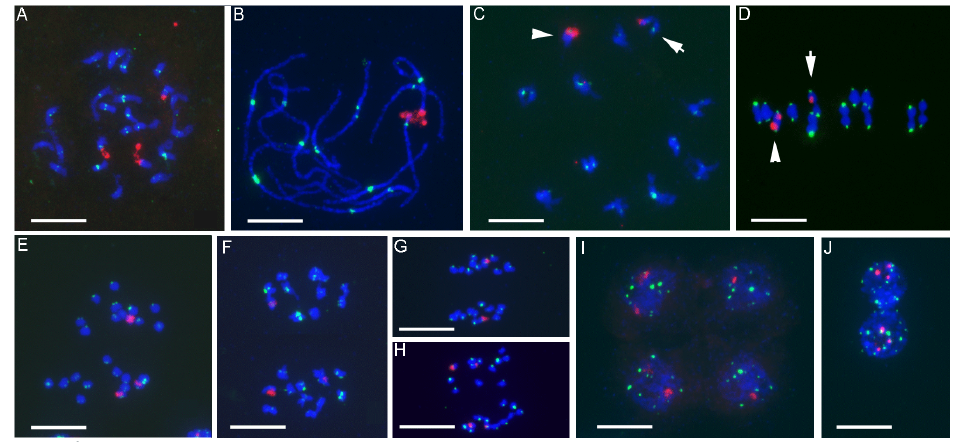
Fig. 2. Mitotic chromosomes and meiotic chromosome behaviors of IR64 × Ol (2n = 24, AA genome).A, Mitotic metaphase. B, Pachytene. C, Diakinesis. D, Metaphase I. E, Anaphase I. F, Metaphase II. G and H, Anaphase II. I, Tetrad. J, Somatic interphase nuclei. In C and D, arrow indicates the bivalent with a single 45S rDNA signal (heteromorphic 45S rDNA site). Arrowhead indicates the bivalent with two 45S rDNA signals. 45S rDNA and centromeric repeats are indicated in red and green, respectively. Scale bars, 10 µm.
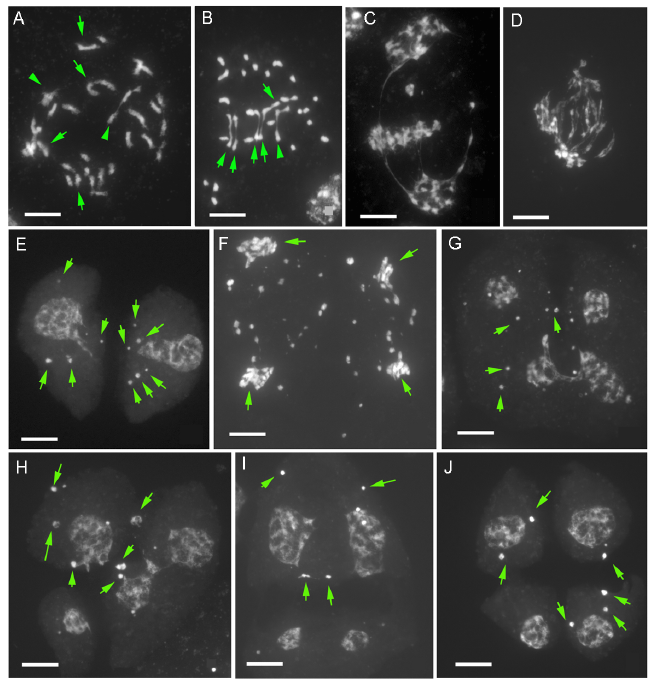
Fig. 3. Meiotic irregularities in pollen mother cells of O. australiensis × O. ridleyi (Oa × Or).A, Diakinesis showing mostly univalents and a low frequency of bivalents (Arrow) and trivalents (Arrowhead). B, Metaphase I showing 23 univalents, 5 bivalents (Arrow) and trivalent (Arrowhead). C and D, Metaphase I to anaphase I showing irregular separation of chromosomes with some chromosomes aggregated at the metaphase plate and some pulled in opposite directions. E, Dyad nuclei with several chromosomes not being included (Arrow). F, Anaphase II showing a high variable number of chromosomes in each pole (Arrow) and many lagging chromosomes.G-J, Tetrad with four microspores having nuclei in varied sizes and several micronuclei (Arrow). Scale bars, 10 µm.
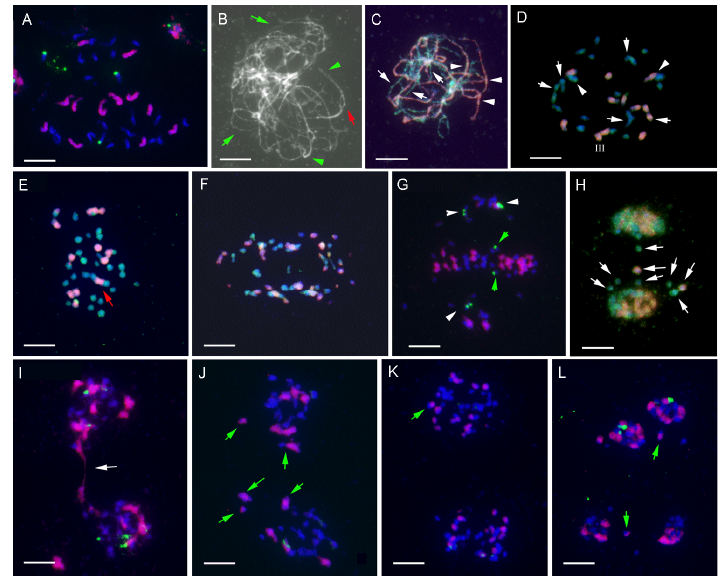
Fig. 4. GISH and 45S rDNA-FISH analysis of meiotic chromosome pairings in O. australiensis × O. ridleyi (Oa × Or).A, Mitotic metaphase: GISH distinguishing chromosomes of the EE genome (Red) from those of the HHJJ genome (Blue). Four 45S rDNA sites (Green) were detected, one from the EE genome and three from the HHJJ genome. B, Pachytene: Showing mostly univalents (Arrow) and a few closely paired bivalents (Arrowhead). Red arrow indicates an unpaired region of a bivalent. C, Pachytene: GISH indicating allosyndetic pairing (Arrowhead) between the EE genome (Red) and HHJJ genome (Blue). Arrows indicate some univalent. D, Diakinesis: GISH showing univalents, bivalents and trivalents. Arrows indicate autosyndetic bivalents, and arrowheads indicate allosyndetic bivalents and trivalents. EE genome and HHJJ genome are indicated in red and blue, respectively. E, Metaphase I: GISH revealing three types of chromosomal associations [i.e., Oa-Oa (Green-Green), Oa-Or (Green-Red), and Or-Or (Red-Red)]. EE genome and HHJJ genome are indicatd in red and blue, while trivalent is indicated by arrow, respectively. F, Anaphase I: GISH showing irregular and unequal allocation of chromosomes at opposite poles. EE genome and HHJJ genome are indicated in red and blue, respectively. G, Metaphase-anaphase I: GISH signals and 45S rDNA FISH signals showing most chromosomes aggregated at a metaphase plate, a few chromosomes moved to opposite poles (Arrowheads), and precocious separation of chromatids (Arrows). EE genome and 45S rDNA are indicated in red and green, respectively. H, Telophase I: GISH showing two daughter nuclei with different chromosome composition and different origin of laggards (Arrow). EE genome and HHJJ genome are indicated in red and green, respectively. I, Telophase I: GISH and FISH showing irregular and unequal allocation of chromosomes at opposite poles. Arrow indicates the chromosome bridge. EE genome and 45S rDNA are indicated in red and green, respectively. J, Metaphase II: GISH and FISH showing different chromosome number and composition at each metaphase plate. Arrows indicate recombinant chromosomes. EE genome and 45S rDNA are indicated in red and green, respectively. K, Anaphase II: GISH and FISH indicating chromatid segregation and different chromosome number and composition at each pole. Arrows indicate recombinant chromosomes. EE genome and 45S rDNA are indicated in red and green, respectively. L, Telophase II: GISH and FISH indicating different number and composition of chromosomes in each pole and all with peripheral micronuclei. Arrows indicate recombinant chromosomes. EE genome and 45S rDNA are indicated in red and green, respectively. FISH, Fluorescence in situ hybridization; GISH, Genomic in situ hybridization. Scale bars, 10 µm.
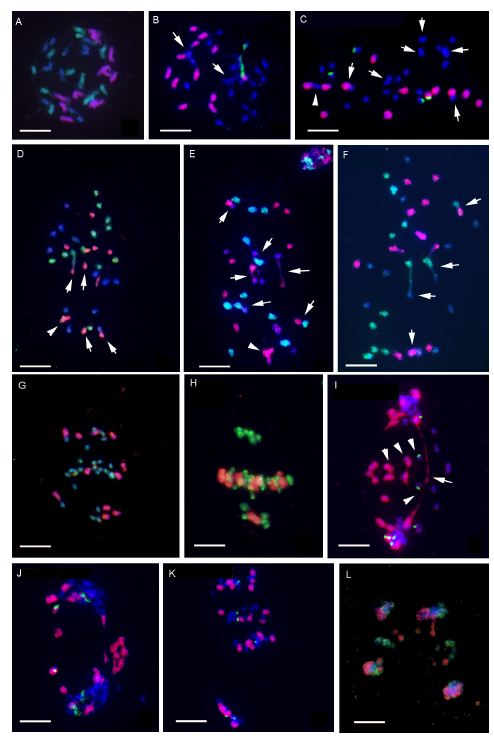
Fig. 5. GISH and 45S rDNA-FISH analysis of meiotic chromosome pairings in O. minuta × O. australiensis (Om × Oa; 2n = 36, BCE genome). A, Mitotic metaphase: GISH signals showing 12 chromosomes from the EE genome (Red) and 12 from the CC genome (Green), and the remaining chromosomes from the BB genome (Blue). B, Diakinesis: GISH and 45S rDNA-FISH signals indicating mostly univalents, four with 45S rDNA sites (Green) including one from the EE genome (Red) and the others from the BBCC genome (Blue). Arrow indicates bivalent. C, Metaphase I: GISH and 45S rDNA-FISH signals indicating mostly univalents scattered irregularly throughout the cell. Arrow indicates bivalent and arrowheads indicate trivalent. EE genome and 45S rDNA are indicated in red and green, respectively. D-F, Metaphase I: GISH signals demonstrating allosyndetic (Arrows) and autosyndetic (Arrowhead) pairing. Green signals indicate chromosomes of CC genome; red signals indicate BB genome in D and EE genome in E and F. G-I, Metaphase I to anaphase I: GISH and 45S rDNA- FISH showing uneven allocation, lagging chromosomes, bridge (Arrow) and precociously separated chromatids (Arrowhead). Red signals indicate chromosomes from the EE genome and green signals from the BBCC genome in G and H and 45S rDNA sites in I. J-L, GISH and 45S rDNA-FISH showing highly meiotic irregularities, including laggards and uneven allocation of chromosomes. J, Telophase I; K, Metaphase II; L, Telophase II. Red signals indicate chromosomes from the EE genome, green signals from 45S rDNA sites in J and K, and CC genome in L. FISH, Fluorescence in situ hybridization; GISH, Genomic in situ hybridization. Scale bars, 10 µm.
| Hybrid | Univalent | Bivalent | Trivalent |
|---|---|---|---|
| Oa × Or | 24.30 ± 3.88 (17-30) | 4.40 ± 1.63 (2-8) | 0.97 ± 1.20 (0-4) |
| Om × Oa | 20.67 ± 3.75 (14-27) | 4.37 ± 1.75 (1-8) | 2.20 ± 1.19 (0-5) |
Table 1 Chromosome associations in pollen mother cells (PMCs) at diakinesis in hybrids O. australiensis (Oa) × O. ridleyi (Or) and O. minuta (Om).
| Hybrid | Univalent | Bivalent | Trivalent |
|---|---|---|---|
| Oa × Or | 24.30 ± 3.88 (17-30) | 4.40 ± 1.63 (2-8) | 0.97 ± 1.20 (0-4) |
| Om × Oa | 20.67 ± 3.75 (14-27) | 4.37 ± 1.75 (1-8) | 2.20 ± 1.19 (0-5) |
| Hybrid/parental species | Genome | Chromosome number | Accession |
|---|---|---|---|
| IR64 × O. longistaminata | AA | 2n = 24 | |
| O. australiensis × O. ridleyi | EHJ | 3n = 36 | |
| O. minuta × O. australiensis | BCE | 3n = 36 | |
| O. sativa | AA | 2n = 24 | IR64 |
| O. longistaminata | AA | 2n = 24 | 110404 |
| O. punctate a | BB | 2n = 24 | 105690 |
| O. officinalis a | CC | 2n = 24 | 100896 |
| O. minuta | BBCC | 2n = 48 | 101141 |
| O. australiensis | EE | 2n = 24 | 105270 |
| O. ridleyi | HHJJ | 2n = 48 | 100821 |
Table 2 Chromosome number and genomic composition of interspecific hybrids and their parental species.
| Hybrid/parental species | Genome | Chromosome number | Accession |
|---|---|---|---|
| IR64 × O. longistaminata | AA | 2n = 24 | |
| O. australiensis × O. ridleyi | EHJ | 3n = 36 | |
| O. minuta × O. australiensis | BCE | 3n = 36 | |
| O. sativa | AA | 2n = 24 | IR64 |
| O. longistaminata | AA | 2n = 24 | 110404 |
| O. punctate a | BB | 2n = 24 | 105690 |
| O. officinalis a | CC | 2n = 24 | 100896 |
| O. minuta | BBCC | 2n = 48 | 101141 |
| O. australiensis | EE | 2n = 24 | 105270 |
| O. ridleyi | HHJJ | 2n = 48 | 100821 |
| [1] | Abbasi F M, Brar D S, Carpena A L, Fukui K, Khush G S. 1999. Detection of autogenetic and allosyndetic pairing among A and E genomes of Oryza through genomic in situ hybridization. Rice Genet Newsl, 16: 24-25. |
| [2] | Abbasi F M, Ahmad H, Sajid M, Inamullah, Brar D S. 2009. Detecting nature of chromosome pairing in A and E genomes of Oryza. Cytologia, 74: 229-233. |
| [3] | Abbasi F M, Shah A H, Perveen F, Afzal M, Sajid M, Masood R, Nawaz F. 2010. Genomic affinity between Oryza sativa and Oryza brachyantha as revealed by in situ hybridization and chromosome pairing. Afr J Biotechnol, 9: 3068-3072. |
| [4] | Amante-Bordeos A, Sitch L A, Nelson R, Damacio R D, Oliva N P, Aswidinnoor H, Leung H. 1992. Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice, Oryza sativa. Theor Appl Genet, 84: 345-354. |
| [5] | Ammiraju J S S, Luo M Z, Goicoechea J L, Wang W M, Kudrna D, Mueller C, Talag J, Kim H, Sisneros N B, Blackmon B, Fang E, Tomkins J B, Brar D, MacKill D, McCouch S, Kurata N, Lambert G, Galbraith D W, Arumuganathan K, Rao K, Walling J G, Gill N, Yu Y, San Miguel P, Soderlund C, Jackson S, Wing R A. 2006. The Oryza bacterial artificial chromosome library resource: Construction and analysis of 12 deep coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res, 16: 140-147. |
| [6] | Anamthawat-Jonsson K, Schwarzacher T, Leitch A R, Bennett M D, Heslop-Harrison J S. 1990. Discrimination between closely related Triticeae species using genomic DNA as a probe. Theor Appl Genet, 79(6): 721-728. |
| [7] | Anamthawat-Jonsson K, Reader S M. 1995. Preannealing of total genomic DNA probes for simultaneous genomic in situ hybridization. Genome, 38(8): 814-816. |
| [8] | Angeles-Shim R B, Vinarao R B, Marathi B, Jena K K. 2014. Molecular analysis of Oryza latifolia Desv. (CCDD genome)- derived introgression lines and identification of value-added traits for rice (O. sativa L.) improvement. J Hered, 105(5): 676-689. |
| [9] | Baghyalakshmi K, Jeyaprakash P, Ramchander S, Radhamani T, Raveendran M. 2018. Comparative study on backcross inbred lines of IR64 rice (Oryza sativa L.) introgressed with drought QTLs under varied moisture regimes over different seasons. Int J Curr Microbiol App Sci, 7(1): 2716-2725. |
| [10] | Brar D S, Elloran R, Khush G S. 1991. Interspecific hybrids produced through embryo rescue between cultivated and eight wild species of rice. Rice Genet Newsl, 8: 91-93. |
| [11] | Brar D S, Khush G S. 1995. Wide hybridization for enhancing resistance to biotic and abiotic stresses in rainfed lowland rice. In: Fragile Lives in Fragile Ecosystems. Manila, the Philippines: International Rice Research Institute: 901-910. |
| [12] | Brar D S, Khush G S. 1997. Alien introgression in rice. Plant Mol Biol, 35: 35-47. |
| [13] | Chang K D, Fang S A, Chang F C, Chung M C. 2010. Chromosomal conservation and sequence diversity of ribosomal RNA genes of two distant Oryza species. Genomics, 96: 181-190. |
| [14] | Chen C B, Xu Y Y, Ma H, Chong K. 2005. Cell biological characterization of male meiosis and pollen development in rice. J Integr Plant Biol, 47: 734-744. |
| [15] | Chung M C, Lee Y I, Cheng Y Y, Chou Y J, Lu C F. 2008. Chromosomal polymorphism of ribosomal genes in the genusOryza. Theor Appl Genet, 116: 745-753. |
| [16] | Chung M C.2015. Chromosome techniques and FISH. In: Yeung E C T, Stasolla C, Summer M J, Huang B Q. Plant microtechniques and protocols. Switzerland: Springer International Publishing: 287-309. |
| [17] | Dawe R K. 1998. Meiotic chromosome organization and segregation in plants. Annu Rev Plant Physiol Plant Mol Biol, 49: 371-395. |
| [18] | Dong F, Miller J T, Jackson S A, Wang G L, Ronald P C, Jiang J. 1998. Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci USA, 95: 8135-8140. |
| [19] | Dupl'áková N, Dobrev P I, Reňák D, Honys D. 2016. Rapid separation of Arabidopsis male gametophyte developmental stages using a Percoll gradient. Nat Protoc, 11: 1817-1832. |
| [20] | Hue N T N, Ram T, Barrion A A, Brar D S. 2003. Characterization of monosomic alien addition lines of Oryza ridleyi through genomic in situ hybridization using meiotic chromosomes. Rice Genet Newsl, 20: 110-111. |
| [21] | Ikehashi H, Araki H. 2008. Genetics of F1 sterility in remote crosses of rice. In: Banta S. Rice Genetics I. Manila, the Philippines: IRRI: 119-130. |
| [22] | Iwata A, Gao D Y, Ohmido N, Jackson S A. 2014. Molecular cytogenetics of rice and its wild relatives. In: Zhang Q F, Wing R A. Genetics and Genomics of Rice: Plant Genetics and Genomics: Crops and Models. New York, USA: Springer: 71-79. |
| [23] | Jauhar P P, Joppa L R. 1996. Chromosome pairing as a tool in genome analysis: Merits and limitations. In: Jauhar P P. Methods of Genome Analysis in Plants. Boca Raton, FL: CRC Press: 9-37. |
| [24] | Jena K K, Khush G S. 1984. Embryo rescue of interspecific hybrids and its scope in rice improvement. Rice Genet Newsl, 1: 133-134. |
| [25] | Jena K K, Khush G S. 1989. Monosomic alien addition lines of rice: Production, morphology, cytology, and breeding behavior. Genome, 32(3): 449-455. |
| [26] | Jena K K, Multani D S, Khush G S. 1991. Monosomic alien addition lines of Oryza australiensis and alien gene transfer. In: Rice Genetics. II. Manila, the Philippines: International Rice Research Institute: 728. |
| [27] | Jena K K, Khush G S, Kochert G. 1992. RFLP analysis of rice (Oryza sativa L.) introgression lines. Theor Appl Genet, 84: 608-616. |
| [28] | Jena K K. 2010. The species of the genus Oryza and transfer of useful genes from wild species into cultivated rice, O. sativa. Breeding Sci, 60(5): 518-523. |
| [29] | Kalinka A, Achrem M, Rogalska S M. 2010. Cytomixis-like chromosomes/chromatin elimination from pollen mother cells (PMCs) in wheat-rye allopolyploids. Nucleus, 53: 69-83. |
| [30] | Kalloo G. 1992. Utilization of wild species. In: Kalloo G, Chowdhury J B. Distant Hybridization of Crop Plants. Berlin, Heidelberg, Germany: Springer: 149-167. |
| [31] | Khush G S, Brar D S. 1988. Wide hybridization in plant breeding. In: Zakri A H. Plant Breeding and Genetic Engineering. SABRAO, Malaysia: 141-188. |
| [32] | Khush G S, Bacalangco E, Ogawa T. 1990. A new gene for resist to bacterial blight from O. longistaminata. Rice Genet Newsl, 7: 121-122. |
| [33] | Kopecký D, Lukaszewski A J, Doležel J. 2008. Cytogenetics of Festulolium (Festuca × Lolium hybrids). Cytogenet Genome Res, 120: 370-383. |
| [34] | Le H T, Armstrong K C, Miki B. 1989. Detection of rye DNA in wheat-rye hybrids and wheat translocation stocks using total genomic DNA as a probe. Plant Mol Biol Rep, 7: 150-158. |
| [35] | Lee Y I, Chang F C, Chung M C. 2011. Chromosome pairing affinities in interspecific hybrids reflect phylogenetic distances among lady’s slipper orchids (Paphiopedilum). Ann Bot, 108(1): 113-121. |
| [36] | Lu B R, Naredo M E B, Juliano A B, Jackson M T. 1998. Taxonomic status of Oryza glumaepatula Steud. III. Assessment of genomic affinity among AA genome species from the New World, Asia, and Australia. Genet Res Crop Evol, 45: 215-223. |
| [37] | Mariam A L, Zakri A H, Mahani M C, Normah M N. 1996. Interspecific hybridization of cultivated rice,Oryza sativa L. with the wild rice, O. minuta Presl. Theor Appl Genet, 93: 664-671. |
| [38] | Mason A S, Huteau V, Eber F, Coriton O, Yan G J, Nelson M N, Cowling W A, Chèvre A M. 2010. Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCAB Brassica interspecific hybrids. Chrom Res, 18(6): 655-666. |
| [39] | Multani D S, Jena K K, Brar D S, de los Reyes B G, Angeles E R, Khush G S. 1994. Development of monosomic alien addition lines and introgression of genes from Oryza australiensis Domin. to cultivated rice O. sativa L. Theor Appl Genet, 88: 102-109. |
| [40] | Oka H I. 1974. Analysis of genes controlling F1 sterility in rice by the use of isogenic lines. Genetics, 77(3): 521-534. |
| [41] | Ouyang Y D, Liu Y G, Zhang Q F. 2010. Hybrid sterility in plant: Stories from rice. Curr Opin Plant Biol, 13(2): 186-192. |
| [42] | Piegu B, Guyot R, Picault N, Roulin A, Sanyal A, Kim H, Collura K, Bra D S, Jackson S, Wing R A, Panaud O. 2006. Doubling genome size without polyploidization: Dynamics of retrotransposition- driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res, 16(10): 1262-1269. |
| [43] | Reddi T V V S, Rao D R M. 2000. Cytology of induced desynaptic mutants in rice. Cytologia, 65: 35-41. |
| [44] | Sanchez P L, Wing R A, Brar D S. 2014. The wild relative of rice: Genomes and genomics. In: Zhang Q F, Wing R A. Genetics and Genomics of Rice, Plant Genetics and Genomics: Crops and Models. New York, USA: Springer: 9-25. |
| [45] | Schwarzacher T, Leitch A R, Bennett M D, Heslop-Harrison J S. 1989. In situ localization of parental genomes in a wide hybrid. Ann Bot, 64: 315-324. |
| [46] | Sitch L A, Dalmacio R D, Romero G O. 1989. Crossability of wild Oryza species and their potential use for improvement of cultivated rice. Rice Genet Newsl, 6: 58-60. |
| [47] | Sitch L A. 1990. Incompatibility barriers operating in crosses of Oryza sativa with related species and genera. In: Gustafson J P. Genetic Manipulation in Plant Improvement: II. New York, USA: Plenum Press: 77-93. |
| [48] | Stein J C, Yu Y, Copetti D, Zwickl D J, Zhang L, Zhang C J, Chougule K, Gao D Y, Iwata A, Goicoechea J L, Wei S R, Wang J, Liao Y, Wang M H, Jacquemin J, Becker C, Kudrna D, Zhang J W, Londono C E M, Song X, Lee S, Sanchez P, Zuccolo A, Ammiraju J S S, Talag J, Danowitz A, Rivera L F, Gschwend A R, Noutsos C, Wu C C, Kao S M, Zeng J W, Wei F J, Zhao Q, Feng Q, Baidouri M E, Carpentier M C, Lasserre E, Cooke R, da Rosa Farias D, da Maia L C, dos Santos R S, Nyberg K G, McNally K L, Mauleon R, Alexandrov N, Schmutz J, Flowers D, Fan C Z, Weigel D, Jena K K, Wicker T, Chen M S, Han B, Henry R, Hsing Y C, Kurata N, de Oliveira A C, Panaud O, Jackson S A, Machado C A, Sanderson M J, Long M Y, Ware D, Wing R A. 2018. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet, 50: 285-296. |
| [49] | Tikapunya T, Fox G P, Furtado A, Henry R J. 2016. Grain physical characteristic of the Australian wild rice. Plant Genet Res, 15: 1-12. |
| [50] | Tikapunya T, Zou W, Yu W W, Powell P O, Fox G P, Furtado A, Henry R J, Gilbert R G. 2017. Molecular structures and properties of starch of Australian wild rice. Carb Polym, 172: 213-222. |
| [51] | Uozu S, Ikehashi H, Ohmido N, Ohtsubo H, Ohtsubo E, Fukui K. 1997. Repetitive sequences: Cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol Biol, 35: 791-799. |
| [52] | Vaughan D A. 1994. Wild Relatives of Rice: Genetic Resources Handbook. Los Banos, the Philippines: International Rice Research Institute. |
| [53] | Xie Q Z, Kang H N, Sparkes D L, Tao S T, Fan X B, Xu L H, Fan X Y, Sha L, Zhang H, Wang Y W, Zeng J, Zhou Y C. 2013. Mitotic and meiotic behavior of rye chromosomes in wheat: Psathyrostachys huashanica amphiploid × Triticale progeny. Genet Mol Res, 12(3): 2537-2548. |
| [54] | Xiong Z Y, Tan G X, He G Y, He G C, Song Y C. 2006. Cytogenetic comparisons between A and G genomes in Oryza using genomic in situ hybridization. Cell Res, 16: 260-266. |
| [55] | Yan H H, Min S K, Zhu L H. 1999. Visualization of Oryza eichingeri chromosomes in intergenomic hybrid plants from O. sativa × O. eichingeri via fluorescent in situ hybridization. Genome, 42(1): 48-51. |
| [56] | Zhao Z G, Zhu S S, Zhang Y H, Bian X F, Wang Y, Jiang L, Liu X, Chen L M, Liu S J, Zhang W W, Ikehashi H, Wan J M. 2011. Molecular analysis of an additional case of hybrid sterility in rice (Oryza sativa L.). Planta, 233: 485-494. |
| [1] | Shalini Pulipati, Suji Somasundaram, Nitika Rana, Kavitha Kumaresan, Mohamed Shafi, Peter Civáň, Gothandapani Sellamuthu, Deepa Jaganathan, Prasanna Venkatesan Ramaravi, S. Punitha, Kalaimani Raju, Shrikant S. Mantri, R. Sowdhamini, Ajay Parida, Gayatri Venkataraman. Diversity of Sodium Transporter HKT1;5 in Genus Oryza [J]. Rice Science, 2022, 29(1): 31-46. |
| [2] | Tao Wang, Lijuan Lou, Zeyu Li, Lianguang Shang, Quan Wang. Cloning and Characterization of Protein Prenyltransferase Alpha Subunit in Rice [J]. Rice Science, 2021, 28(6): 557-566. |
| [3] | Elisa B. Gerona Maria, P. Deocampo Marjorie, A. Egdane James, M. Ismail Abdelbagi, L. Dionisio-Sese Maribel. Physiological Responses of Contrasting Rice Genotypes to Salt Stress at Reproductive Stage [J]. Rice Science, 2019, 26(4): 207-219. |
| [4] | Haritha G., Sudhakar T., Chandra D., Ram T., Divya B., Sarla N.. Informative ISSR Markers Help Identify Genetically Distinct Accessions of Oryza rufipogon in Yield Improvement [J]. Rice Science, 2016, 23(5): 225-241. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||