
Rice Science ›› 2016, Vol. 23 ›› Issue (5): 225-241.DOI: 10.1016/j.rsci.2016.08.001
• Orginal Article • Next Articles
Haritha G., Sudhakar T., Chandra D., Ram T., Divya B., Sarla N.( )
)
Received:2015-10-09
Accepted:2016-02-01
Online:2016-09-05
Published:2016-06-12
Haritha G., Sudhakar T., Chandra D., Ram T., Divya B., Sarla N.. Informative ISSR Markers Help Identify Genetically Distinct Accessions of Oryza rufipogon in Yield Improvement[J]. Rice Science, 2016, 23(5): 225-241.
Add to citation manager EndNote|Ris|BibTeX
| N | Acc No. | Species | Source | N | Acc No. | Species | Source | N | Acc No. | Species | Source |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 106147 | O. rufipogon | Laos | 31 | 81998 | O. rufipogon | Papua new guinea | 61 | WR119 | O. rufipogon | India |
| 2 | 106277 | O. rufipogon | Papua new guinea | 32 | 105125 | O. rufipogon | Papua new guinea | 62 | WR120 | O. rufipogon | India |
| 3 | 106507 | O. rufipogon | Myanmar | 33 | 106162 | O. rufipogon | Laos | 63 | 81879 | O. rufipogon | India |
| 4 | 106462 | O. rufipogon | India | 34 | 81877 | O. rufipogon | India | 64 | 81894 | O. rufipogon | India |
| 5 | 106131 | O. rufipogon | India | 35 | 81802 | O. rufipogon | Indonesia | 65 | 86476 | O. rufipogon | India |
| 6 | 106459 | O. rufipogon | India | 36 | 80712 | O. rufipogon | India | 66 | 102166 | O. nivara | India |
| 7 | 106427 | O. rufipogon | Vietnam | 37 | 81878 | O. rufipogon | India | 67 | 105320 | O. nivara | India |
| 8 | 106453 | O. rufipogon | Indonesia | 38 | 106274 | O. rufipogon | Papua new guinea | 68 | 105333 | O. nivara | India |
| 9 | 106123 | O. rufipogon | India | 39 | 104640 | O. rufipogon | Vietnam | 69 | 106052 | O. nivara | India |
| 10 | 106437 | O. rufipogon | Vietnam | 40 | 81977 | O. rufipogon | Indonesia | 70 | 106495 | O. nivara | India |
| 11 | 106338 | O. rufipogon | Cambodia | 41 | 104425 | O. rufipogon | Thailand | 71 | 105809 | O. nivara | Thailand |
| 12 | 106122 | O. rufipogon | India | 42 | 104423 | O. rufipogon | Thailand | 72 | 81848 | O. nivara | India |
| 13 | 106271 | O. rufipogon | Papua new guinea | 43 | 80669 | O. rufipogon | India | 73 | 104721 | O. nivara | Thailand |
| 14 | 81890 | O. rufipogon | India | 44 | 80762 | O. rufipogon | Myanmar | 74 | 105894 | O. nivara | Bangladesh |
| 15 | 81892 | O. rufipogon | India | 45 | 81986 | O. rufipogon | Cambodia | 75 | 101230 | O. longistaminata | Sierra Leone |
| 16 | 81894 | O. rufipogon | India | 46 | 81987 | O. rufipogon | Cambodia | 76 | 101207 | O. longistaminata | Ivory coast |
| 17 | 81884 | O. rufipogon | India | 47 | 81989 | O. rufipogon | Myanmar | 77 | IR64 | O. sativa | IRRI |
| 18 | 106465 | O. rufipogon | Laos | 48 | 80592 | O. rufipogon | India | 78 | Swarna | O. sativa | India |
| 19 | 106515 | O. rufipogon | Vietnam | 49 | 81880 | O. rufipogon | India | 79 | Rasi | O. sativa | India |
| 20 | 106119 | O. rufipogon | India | 50 | 80781 | O. rufipogon | India | 80 | 9314 | O. sativa | China |
| 21 | 106125 | O. rufipogon | India | 51 | 81895 | O. rufipogon | India | 81 | BSI115 | O. sativa | Indonesia |
| 22 | 80671 | O. rufipogon | India | 52 | 106118 | O. rufipogon | India | 82 | Moroberekan | O. sativa | Guinea |
| 23 | 106144 | O. rufipogon | India | 53 | 106115 | O. rufipogon | India | 83 | Nipponbare | O. sativa | Japan |
| 24 | 106407 | O. rufipogon | Vietnam | 54 | 106084 | O. rufipogon | India | 84 | Taipei 309 | O. sativa | Taiwan, China |
| 25 | 82011 | O. rufipogon | India | 55 | 105910 | O. rufipogon | Thailand | 85 | N22 | O. sativa | India |
| 26 | 106163 | O. rufipogon | Laos | 56 | 106057 | O. rufipogon | India | 86 | FR13A | O. sativa | India |
| 27 | 106149 | O. rufipogon | Laos | 57 | 104599 | O. rufipogon | Sri Lanka | 87 | Dular | O. sativa | India |
| 28 | 106340 | O. rufipogon | Myanmar | 58 | 104606 | O. rufipogon | Sri Lanka | 88 | Basmati 370 | O. sativa | India |
| 29 | 81978 | O. rufipogon | Indonesia | 59 | 106083 | O. rufipogon | India | 89 | Type 3 | O. sativa | India |
| 30 | 81990 | O. rufipogon | Myanmar | 60 | 104639 | O. rufipogon | Thailand | 90 | CSR30 | O. sativa | India |
Table 1 List of O. rufipogon, O. nivara, O. longistaminata and O. sativa genotypes used and their origins.
| N | Acc No. | Species | Source | N | Acc No. | Species | Source | N | Acc No. | Species | Source |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 106147 | O. rufipogon | Laos | 31 | 81998 | O. rufipogon | Papua new guinea | 61 | WR119 | O. rufipogon | India |
| 2 | 106277 | O. rufipogon | Papua new guinea | 32 | 105125 | O. rufipogon | Papua new guinea | 62 | WR120 | O. rufipogon | India |
| 3 | 106507 | O. rufipogon | Myanmar | 33 | 106162 | O. rufipogon | Laos | 63 | 81879 | O. rufipogon | India |
| 4 | 106462 | O. rufipogon | India | 34 | 81877 | O. rufipogon | India | 64 | 81894 | O. rufipogon | India |
| 5 | 106131 | O. rufipogon | India | 35 | 81802 | O. rufipogon | Indonesia | 65 | 86476 | O. rufipogon | India |
| 6 | 106459 | O. rufipogon | India | 36 | 80712 | O. rufipogon | India | 66 | 102166 | O. nivara | India |
| 7 | 106427 | O. rufipogon | Vietnam | 37 | 81878 | O. rufipogon | India | 67 | 105320 | O. nivara | India |
| 8 | 106453 | O. rufipogon | Indonesia | 38 | 106274 | O. rufipogon | Papua new guinea | 68 | 105333 | O. nivara | India |
| 9 | 106123 | O. rufipogon | India | 39 | 104640 | O. rufipogon | Vietnam | 69 | 106052 | O. nivara | India |
| 10 | 106437 | O. rufipogon | Vietnam | 40 | 81977 | O. rufipogon | Indonesia | 70 | 106495 | O. nivara | India |
| 11 | 106338 | O. rufipogon | Cambodia | 41 | 104425 | O. rufipogon | Thailand | 71 | 105809 | O. nivara | Thailand |
| 12 | 106122 | O. rufipogon | India | 42 | 104423 | O. rufipogon | Thailand | 72 | 81848 | O. nivara | India |
| 13 | 106271 | O. rufipogon | Papua new guinea | 43 | 80669 | O. rufipogon | India | 73 | 104721 | O. nivara | Thailand |
| 14 | 81890 | O. rufipogon | India | 44 | 80762 | O. rufipogon | Myanmar | 74 | 105894 | O. nivara | Bangladesh |
| 15 | 81892 | O. rufipogon | India | 45 | 81986 | O. rufipogon | Cambodia | 75 | 101230 | O. longistaminata | Sierra Leone |
| 16 | 81894 | O. rufipogon | India | 46 | 81987 | O. rufipogon | Cambodia | 76 | 101207 | O. longistaminata | Ivory coast |
| 17 | 81884 | O. rufipogon | India | 47 | 81989 | O. rufipogon | Myanmar | 77 | IR64 | O. sativa | IRRI |
| 18 | 106465 | O. rufipogon | Laos | 48 | 80592 | O. rufipogon | India | 78 | Swarna | O. sativa | India |
| 19 | 106515 | O. rufipogon | Vietnam | 49 | 81880 | O. rufipogon | India | 79 | Rasi | O. sativa | India |
| 20 | 106119 | O. rufipogon | India | 50 | 80781 | O. rufipogon | India | 80 | 9314 | O. sativa | China |
| 21 | 106125 | O. rufipogon | India | 51 | 81895 | O. rufipogon | India | 81 | BSI115 | O. sativa | Indonesia |
| 22 | 80671 | O. rufipogon | India | 52 | 106118 | O. rufipogon | India | 82 | Moroberekan | O. sativa | Guinea |
| 23 | 106144 | O. rufipogon | India | 53 | 106115 | O. rufipogon | India | 83 | Nipponbare | O. sativa | Japan |
| 24 | 106407 | O. rufipogon | Vietnam | 54 | 106084 | O. rufipogon | India | 84 | Taipei 309 | O. sativa | Taiwan, China |
| 25 | 82011 | O. rufipogon | India | 55 | 105910 | O. rufipogon | Thailand | 85 | N22 | O. sativa | India |
| 26 | 106163 | O. rufipogon | Laos | 56 | 106057 | O. rufipogon | India | 86 | FR13A | O. sativa | India |
| 27 | 106149 | O. rufipogon | Laos | 57 | 104599 | O. rufipogon | Sri Lanka | 87 | Dular | O. sativa | India |
| 28 | 106340 | O. rufipogon | Myanmar | 58 | 104606 | O. rufipogon | Sri Lanka | 88 | Basmati 370 | O. sativa | India |
| 29 | 81978 | O. rufipogon | Indonesia | 59 | 106083 | O. rufipogon | India | 89 | Type 3 | O. sativa | India |
| 30 | 81990 | O. rufipogon | Myanmar | 60 | 104639 | O. rufipogon | Thailand | 90 | CSR30 | O. sativa | India |
| Primer | Sequence repeat motif (5′-3′) | Annealing temperature (°C) |
|---|---|---|
| UBC-807 | (AG)8T | 47 |
| UBC-808 | (AG)8C | 49 |
| UBC-809 | (AG)8G | 49 |
| UBC-810 | (GA)8T | 47 |
| UBC-811 | (GA)8C | 47 |
| UBC-812 | (GA)8A | 47 |
| UBC-823 | (TC)8C | 49 |
| UBC-824 | (GA)8G | 49 |
| UBC-834 | (GA)8YT | 48 |
| UBC-835 | (AG)8TC | 50 |
| UBC-840 | (GA)8YT | 47 |
| UBC-842 | (GA)8YG | 50 |
| UBC-853 | (TC)8RT | 48 |
| UBC-854 | (TC)8RG | 48 |
| UBC-884A | HBH(AG)7 | 47 |
| UBC-884B | ACTGCT(GA)7 | 50 |
| UBC-885 | GCCG(AG)6 | 50 |
Table 2 Details of 17 primers used in ISSR-PCR.
| Primer | Sequence repeat motif (5′-3′) | Annealing temperature (°C) |
|---|---|---|
| UBC-807 | (AG)8T | 47 |
| UBC-808 | (AG)8C | 49 |
| UBC-809 | (AG)8G | 49 |
| UBC-810 | (GA)8T | 47 |
| UBC-811 | (GA)8C | 47 |
| UBC-812 | (GA)8A | 47 |
| UBC-823 | (TC)8C | 49 |
| UBC-824 | (GA)8G | 49 |
| UBC-834 | (GA)8YT | 48 |
| UBC-835 | (AG)8TC | 50 |
| UBC-840 | (GA)8YT | 47 |
| UBC-842 | (GA)8YG | 50 |
| UBC-853 | (TC)8RT | 48 |
| UBC-854 | (TC)8RG | 48 |
| UBC-884A | HBH(AG)7 | 47 |
| UBC-884B | ACTGCT(GA)7 | 50 |
| UBC-885 | GCCG(AG)6 | 50 |
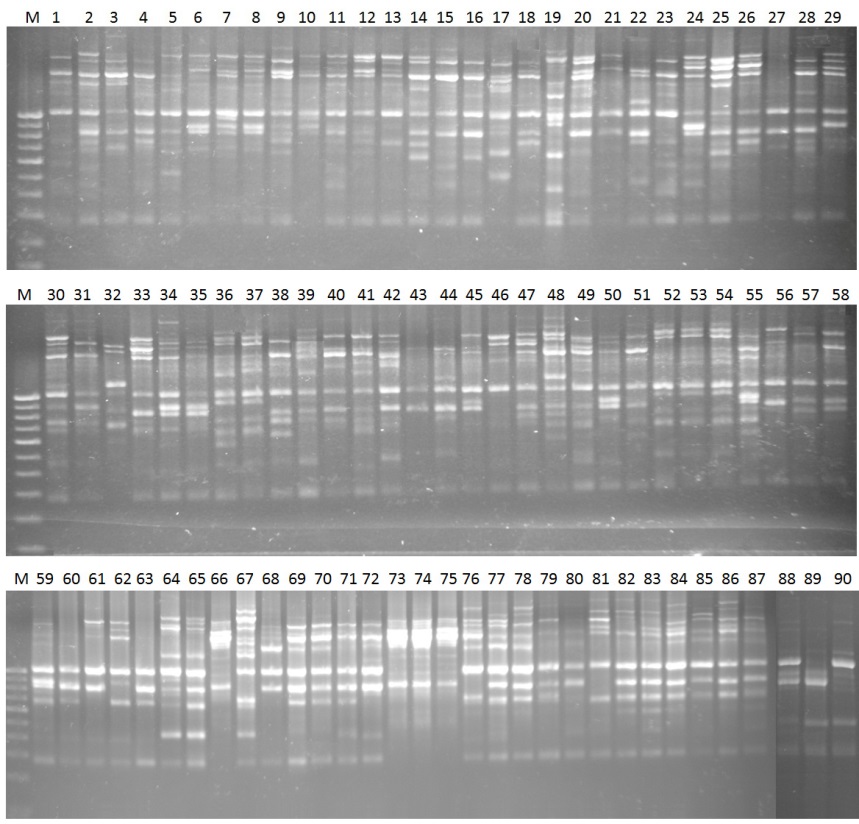
Fig. 1. Fingerprints of 90 samples using primer UBC-823 by inter simple sequence repeat markers. M, 100-bp ladder. 1 to 90 are the numbers of the genotypes which are shown in Table 1.
| Primer | PIC | Resolving power (Rp) | No. of band positions | Total No. of bands amplified | Range of bands amplified by primer b |
|---|---|---|---|---|---|
| UBC-807 | 0.84 | 14.11 | 19 | 635 | 11-53 |
| UBC-808 | 0.7 | 13.27 | 13 | 597 | 12-67 |
| UBC-809 a | 0.65 | 23.53 | 21 | 1059 | 23-83 |
| UBC-810 | 0.59 | 8.69 | 7 | 391 | 41-76 |
| UBC-811 | 0.44 | 10.36 | 7 | 466 | 52-86 |
| UBC-812 a | 0.64 | 19.98 | 18 | 899 | 18-84 |
| UBC-823 | 0.73 | 14.58 | 15 | 656 | 23-79 |
| UBC-824 | 0.72 | 16.27 | 16 | 732 | 26-70 |
| UBC-834 a | 0.69 | 17.09 | 17 | 769 | 15-81 |
| UBC-835 | 0.84 | 8.69 | 14 | 391 | 4-81 |
| UBC-840 a | 0.67 | 18.04 | 17 | 812 | 12-70 |
| UBC-842 a | 0.61 | 18.6 | 16 | 837 | 11-86 |
| UBC-853 | 0.84 | 12.93 | 18 | 582 | 4-59 |
| UBC-854 | 0.64 | 14.93 | 13 | 672 | 22-85 |
| UBC-884A | 0.62 | 16.18 | 14 | 728 | 17-80 |
| UBC-884B | 0.79 | 9.51 | 12 | 428 | 5-82 |
| UBC-885 a | 0.64 | 18 | 16 | 810 | 21-80 |
Table 3 Details for primers.
| Primer | PIC | Resolving power (Rp) | No. of band positions | Total No. of bands amplified | Range of bands amplified by primer b |
|---|---|---|---|---|---|
| UBC-807 | 0.84 | 14.11 | 19 | 635 | 11-53 |
| UBC-808 | 0.7 | 13.27 | 13 | 597 | 12-67 |
| UBC-809 a | 0.65 | 23.53 | 21 | 1059 | 23-83 |
| UBC-810 | 0.59 | 8.69 | 7 | 391 | 41-76 |
| UBC-811 | 0.44 | 10.36 | 7 | 466 | 52-86 |
| UBC-812 a | 0.64 | 19.98 | 18 | 899 | 18-84 |
| UBC-823 | 0.73 | 14.58 | 15 | 656 | 23-79 |
| UBC-824 | 0.72 | 16.27 | 16 | 732 | 26-70 |
| UBC-834 a | 0.69 | 17.09 | 17 | 769 | 15-81 |
| UBC-835 | 0.84 | 8.69 | 14 | 391 | 4-81 |
| UBC-840 a | 0.67 | 18.04 | 17 | 812 | 12-70 |
| UBC-842 a | 0.61 | 18.6 | 16 | 837 | 11-86 |
| UBC-853 | 0.84 | 12.93 | 18 | 582 | 4-59 |
| UBC-854 | 0.64 | 14.93 | 13 | 672 | 22-85 |
| UBC-884A | 0.62 | 16.18 | 14 | 728 | 17-80 |
| UBC-884B | 0.79 | 9.51 | 12 | 428 | 5-82 |
| UBC-885 a | 0.64 | 18 | 16 | 810 | 21-80 |
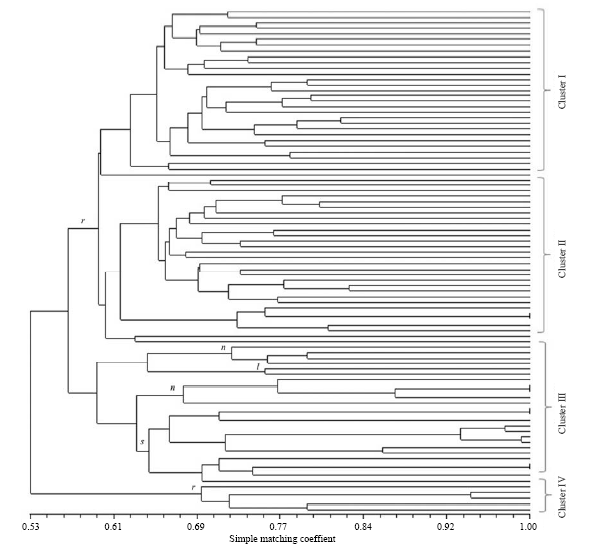
Fig. 4. Un-weighted pair group method with arithmetic mean dendrogram among 90 accessions using 17 primers. r, O. rufipogon; n, O. nivara; l, O. longistaminata; s, O. sativa. Accession numbers (from top to bottom) are 1, 23, 3, 28, 29, 24, 26, 30, 21, 22, 25, 27, 2, 13, 11, 14, 15, 16, 17, 6, 7, 10, 8, 4, 18, 9, 12, 5, 20, 19, 31, 47, 37, 33, 34, 35, 54, 51, 50, 55, 56, 57, 58, 48, 49, 38, 39, 40, 42, 43, 44, 45, 46, 36, 41, 60, 52, 53, 32, 59, 71, 72, 73, 74, 75, 76, 66, 67, 69, 68, 70, 77, 78, 80, 81, 82, 83, 84, 88, 89, 85, 86, 87, 79, 90, 61, 62, 65, 63 and 64.
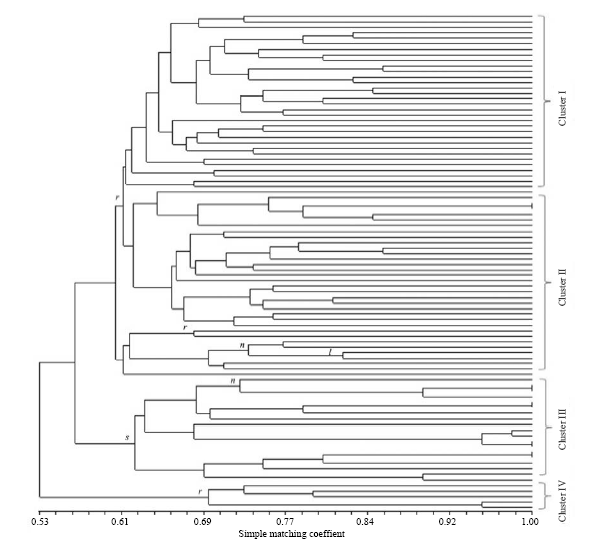
Fig. 5. Un-weighted pair group method with arithmetic mean dendrogram among 90 accessions using 6 informative primers. r, O. rufipogon; n, O. nivara; l, O. longistaminata; s, O. sativa. Accession numbers (from top to bottom) are 1, 23, 27, 2, 13, 11, 4, 18, 21, 6, 8, 7, 10, 9, 12, 14, 15, 16, 22, 20, 24, 30, 28, 29, 26, 57, 17, 25, 5, 32, 3, 58, 19, 36, 41, 60, 52, 53, 38, 31, 49, 33, 34, 35, 51, 37, 54, 48, 50, 39, 40, 42, 43, 44, 45, 46, 47, 55, 56, 71, 74, 75, 76, 72, 73, 59, 66, 67, 69, 68, 77, 78, 85, 80, 70, 81, 82, 83, 84, 86, 87, 79, 90, 88, 89, 61, 63, 64, 62 and 65.
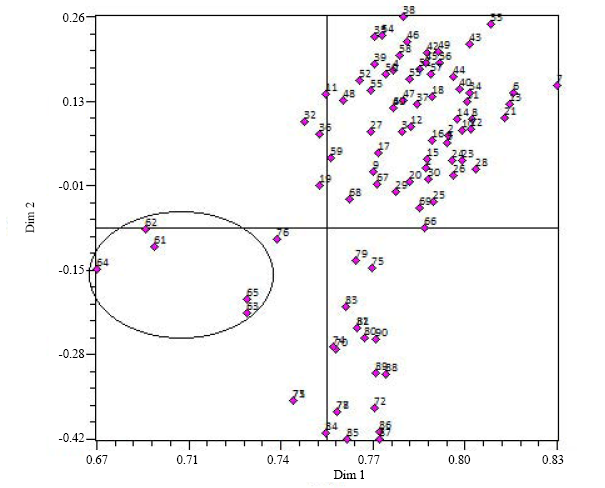
Fig. 6. Two dimensional plots of the genetic relationship among 90 genotypes of Oryza as revealed by principal coordinate analysis (PCoA) in NTSys ver. 2.02. The five O. rufipogon accessions 61, 62, 63, 64 and 65 separated from the other O. rufipogon accessions and grouped together.
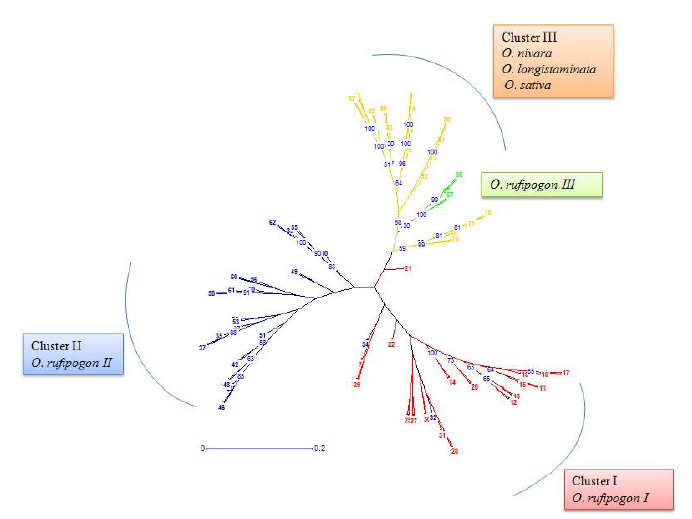
Fig. 7. An unrooted neighbor joining tree showing the genetic relationships among the 90 genotypes. Red indicates 30 O. rufipogon accessions of subgroup I, blue indicates another 30 O. rufipogon accessions of subgroup II, and yellow indicates O. nivara, O. longistaminata and O. sativa accessions. The five distinct O. rufipogon accessions of subgroup III are in green color.
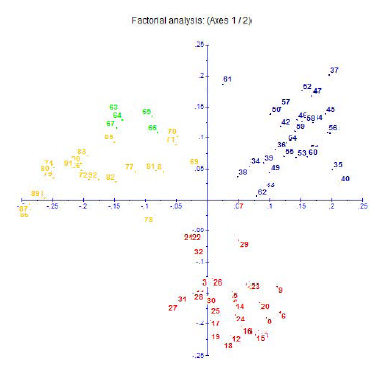
Fig. 8. Factorial correspondence analysis of 90 genotypes using 17 primers. Red indicates O. rufipogon accessions of sub-group I, blue indicates O. rufipogon accessions of sub-group II and yellow indicates O. nivara, O. longistaminata and O. sativa accessions. The five distinct O. rufipogon accessions of sub-group III are in green color.
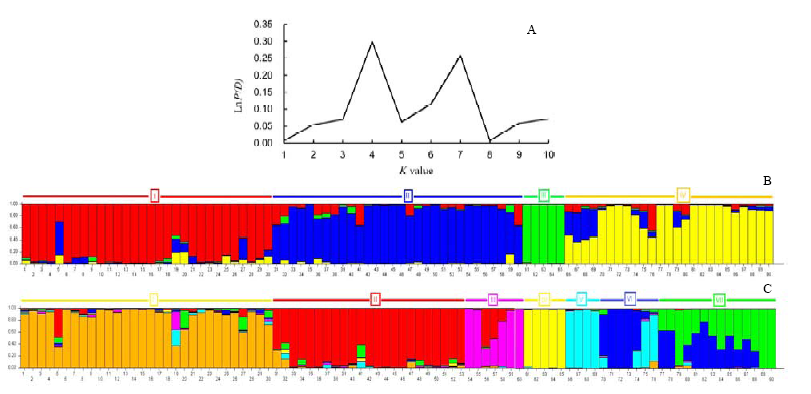
Fig. 9. STRUCTURE analysis of the 90 accessions. A, The graphical representation showing population structure of two peaks. One peak at K = 4 showing largely inter-species differentiation and the other peak at K = 7 showing largely intra-species differentiation within wild and cultivated genotypes. B, A model-based clustering method at K = 4 (the first panel). Each color represents different population groups. I, II, and III represent sub-populations within O. rufipogon; IV represents O. nivara, O. longistaminata and O. sativa. C, A model-based clustering method at K = 7 (the second panel). Each color represents different population groups. I, II, III and IV represent four sub-populations within O. rufipogon; V and VI represent sub-populations within O. nivara accessions and two O. longistaminata also grouped here; VII represents population consisting of all the five sub-groups of O. sativa. A, The graphical representation showing population structure of two peaks. One peak at K = 4 showing largely inter-species differentiation and the other peak at K = 7 showing largely intra-species differentiation within wild and cultivated genotypes. B, A model-based clustering method at K = 4 (the first panel). Each color represents different population groups. I, II, and III represent sub-populations within O. rufipogon; IV represents O. nivara, O. longistaminata and O. sativa. C, A model-based clustering method at K = 7 (the second panel). Each color represents different population groups. I, II, III and IV represent four sub-populations within O. rufipogon; V and VI represent sub-populations within O. nivara accessions and two O. longistaminata also grouped here; VII represents population consisting of all the five sub-groups of O. sativa.
| 1 | Aiello L C.2011. The origins of agriculture: New data, new ideas.Curr Anthropol, 52(S4): S161-S162. |
| 2 | Aggarwal R K, Brar D S, Nandi S, Huang N, Khush G S.1999. Phylogenetic relationships among Oryza species revealed by AFLP markers.Theor Appl Genet, 98: 1320-1328. |
| 3 | Alhasnawi A N, Kadhimi A A, Isahak A, Ashraf M F, Doni F, Mohamad A, Mohtar W, Yusoff W, Radziah C, Zain C M.2015. Application of inter simple sequence repeat (ISSR) for detecting genetic analysis in rice (Oryza sativa L.).J Pure Appl Microbiol, 9(2): 1091-1101. |
| 4 | Ali M L, Sanchez P L, Yu S B, Lorieux M, Eizenga G C.2010. Chromosome segment substitution lines: A powerful tool for the introgression of valuable genes from wild species of rice (Oryza spp.).Rice, 3: 218-234. |
| 5 | Alvarez A, Fuentes J L, Puldon V, Gomez P J, Mora L, Duque M C, Gallego G, Tohme J M.2007. Genetic diversity analysis of Cuban traditional rice (Oryza sativa L.) varieties.Genet Mol Biol, 30: 1109-1117. |
| 6 | Anuradha K, Agarwal S, Batchu A K, Babu A P, Swamy B P M, Longvah T, Sarla N.2012. Evaluating rice germplasm for iron and zinc concentration in brown rice and seed dimensions.J Phytol, 4(1): 19-25. |
| 7 | Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles E R, Qian Q, Kitano H, Matsuoka M.2005. Cytokinin oxidase regulates rice grain production.Science, 29: 741-745. |
| 8 | Barbier P.1989. Genetic variation and ecotypic differentiation in the wild rice species Oryza rufipogon: I. Population differentiation in life-history traits and isozymic loci.Jpn J Genet, 64(4): 259-271. |
| 9 | Barcaccia G.2009. Molecular markers for characterizing and conserving crop plant germplasm. In: Jain S M, Brar D S. Molecular Techniques in Crop Improvement . Dordrecht, Netherlands: Springer: 231-254. |
| 10 | Bimpong I K, Serraj R, Chin J H, Ramos J, Mendoza E M T, Hernandez J E, Mendioro M S, Brar D S.2011. Identification of QTLs for drought related traits in alien introgression lines derived from crosses of rice (Oryza sativa cv. IR64) and O. glaberrima under low land moisture stress.J Plant Biol, 54: 237-250. |
| 11 | Blair M W, Panaud O, McCouch S R.1999. Inter-simple sequence repeats (ISSR) amplification for analysis of microsatellite motif frequency and fingerprinting in rice (Oryza sativa L.).Theor Appl Genet, 98(5): 780-792. |
| 12 | Brar D S, Khush G S.1997. Alien introgression in rice.Plant Mol Biol, 35(1): 35-47. |
| 13 | Brar D S, Khush G S.2002. Transferring genes from wild species into rice. In: Kang M S. Quantitatie Genetics, Genomics and Plant Breeding. Wallingford, UK: CABI: 197-217. |
| 14 | Brar D S, Singh K.2011. Oryza. In: Kole C. Wild Crop Relatives: Genomic and Breeding Resources, Cereals. Berlin, Heidelberg: Springer: 321-365. |
| 15 | Brozynska M, Furtado A, Henry R J.2015. Genomics of crop wild relatives: Expanding the gene pool for crop improvement.Plant Biotechnol J, 14(4): 1070-1085. |
| 16 | Cao Q J, Lu B R, Xia H, Rong J, Sala F, Spada A, Grassi F.2006. Genetic diversity and origin of weedy rice (Oryza sativa f. spontanea) populations found in North Eastern China revealed by simple sequence repeat (SSR) markers.Ann Bot, 98(6): 1241-1252. |
| 17 | Celeste M, Banaticla-Hilario N, van den Berg R G, Hamilton N R S, McNally K L.2013. Local differentiation amidst extensive allele sharing in Oryza nivara and O. rufipogon.Ecol Evol, 3: 3047-3062. |
| 18 | Chen L J, Lee D S, Song Z P, Suh H S, Lu B R.2004. Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives.Ann Bot, 93(1): 67-73. |
| 19 | Choudhary P, Khanna S M, Jain P K, Bharadwaj C, Kumar J, Lakhera P C, Srinivasan R.2013. Molecular characterization of primary gene pool of chickpea based on ISSR markers.Biochem Genet, 51: 306-322. |
| 20 | Davierwala A P, Chowdari K V, Kumar S, Reddy A P K, Ranjekar P K, Gupta V S.2000. Use of three different marker systems to estimate genetic diversity of Indian elite rice varieties.Genetica, 108(3): 269-284. |
| 21 | Evanno G, Regnaut S, Goudet J.2005. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study.Mol Ecol, 14: 2611-2620. |
| 22 | Fu X L, Lu Y G, Liu X D, Li J Q.2008. Progress on transfering elite genes from non-AA genome wild rice into Oryza sativa through interspecific hybridization.Rice Sci, 15(2): 79-87. |
| 23 | Furuta T, Uehara K, Angeles-Shim R B, Shim J, Ashikari M, Takashi T.2014. Development and evaluation of chromosome segment substitution lines (CSSLs) carrying chromosome segments derived from Oryza rufipogon in the genetic background of Oryza sativa L.Breeding Sci, 63(5): 468-475. |
| 24 | Garland S H, Lewin L, Abedinia M, Henry R, Blakeney A.1999. The use of microsatellite polymorphisms for the identification of Australian breeding lines of rice (Oryza sativa L.).Euphytica, 108: 53-63. |
| 25 | Gao H, Williamson S, Bustamante C D.2007. A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data.Genetics, 176(3): 1635-1651. |
| 26 | Garris A J, Tai T H, Coburn J, Kresovich S, McCouch S.2005. Genetic structure and diversity in Oryza sativa L.Genetics, 169(3): 1631-1638. |
| 27 | Girma G, Tesfaye K, Bekele E.2010. Inter simple sequence repeat (ISSR) analysis of wild and cultivated rice species from Ethiopia.Afr J Biotechnol, 9: 5048-5059. |
| 28 | Glaszmann J C.1987. Isozymes and classification of Asian rice varieties.Theor Appl Genet, 74(1): 21-30. |
| 29 | Govindaraj M, Vetriventhan M, Srinivasan M.2015. Importance of genetic diversity assessment in crop plants and its recent advances: An overview of its analytical perspectives.Genet Res Int, 1: 1-14. |
| 30 | He G M, Luo X J, Tian F, Li K G, Zhu Z F, Su W, Qian X Y, Fu Y C, Wang X K, Sun C Q, Yang J S.2006. Haplotype variation in structure and expression of a gene cluster associated with a quantitative trait locus for improved yield in rice.Genom Res, 16(5): 618-626. |
| 31 | Herrera T G, Duque D P, Almeida I P, Nunez T G, Pieters A J, Martinez C P, Tohme J M.2008. Assessment of genetic diversity in Venezuelan rice cultivars using simple sequence repeats markers.Electron J Biotechnol, 11(5): 1-14. |
| 32 | Huang C L, Hwang S Y, Chiang Y C, Lin T P.2008. Molecular evolution of the Pi-ta gene resistant to rice blast in wild rice (Oryza rufipogon).Genetics, 179(3): 1527-1538. |
| 33 | Huang P, Molina J, Flowers J M, Rubinstein S, Jackson S A, Purugganan M D, Schaal B A.2012. Phylogeography of Asian wild rice, Oryza rufipogon: A genome-wide view.Mol Ecol, 21: 4593-4604. |
| 34 | Huang D, Qin G, Liu C, Ma Z F, Zhang Y X, Yan Y.2013. Feasibility of utilization of wild rice (Oryza rufipogon Griff.) genetic diversity in rice breeding for high yield.Adv J Food Sci Technol, 5(5): 640-645. |
| 35 | Jena K K, Khush G S.2000. Exploitation of species in rice improvement: Opportunities, achievements and future challenges. In: Nanda J S. Rice Breeding and Genetic: Research Priorities and Challenges. Endfield, New Hampshire: Science Publisher: 269-284. |
| 36 | Jena S N, Verma S, Nair K N, Srivastava A K, Misra S, Rana T S.2015. Genetic diversity and population structure of the mangrove lime (Merope angulata) in India revealed by AFLP and ISSR markers.Aquat Bot, 120: 260-267. |
| 37 | Jeung J U, Kim B R, Cho Y C, Han S S, Moon H P, Lee Y T, Jena K K.2007. A novel gene, Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice.Theor Appl Genet, 115(8): 1163-1177. |
| 38 | Joshi S P, Gupta V S, Aggarwal R K, Ranjekar P K, Brar D S.2000. Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus Oryza.Theor Appl Genet, 100: 1311-1320. |
| 39 | Juneja S, Das A, Joshi S V, Sharma S, Vikal Y, Patra B C, Bharaj T S, Sidhu J S, Singh K.2006. Oryza nivara (Sharma et Shastry) the progenitor of O. sativa (L.) subspecies indica harbours rich genetic diversity as measured by SSR markers.Curr Sci, 91(8): 1079-1085. |
| 40 | Kaladhar K, Swamy B P M, Babu A P, Reddy C S, Sarla N.2008. Mapping quantitative trait loci for yield traits in BC2F2 population derived from Swarna × O. nivara cross.Rice Genet Newsl, 24: 34-36. |
| 41 | Khan M K, Pandey A, Thomas G, Akkaya M S, Kayis S A, Ozsensoy Y, Hamurcu M, Gezgin S, Topal A, Hakki E E.2015. Genetic diversity and population structure of wheat in India and Turkey. AoB Plants, 7: plv083. |
| 42 | Khshirsagar S S, Samal K C, Rabha M, Bastia D N, Rout G R.2014. Identification of variety diagnostic molecular marker of high yielding rice varieties.Proc Natl Acad Sci Ind: Biol Sci, 84(2): 389-396. |
| 43 | Khush G S, Ling K C, Aquino R C, Aguiero V M.1977. Breeding for resistance to grassy stunt in rice. In: International Seminar of Society for Advancement Breeding Research in Asia and Oceania. Canberra, Australia, 12-13 February, 1(4b): 3-9. |
| 44 | Kumar P N, Sujatha K, Laha G S, Rao K S, Mishra B, Viraktamath B C, Hari Y, Reddy C S, Balachandran S M, Ram T, Madhav M S, Rani N S, Neeraja C N, Reddy G A, Shaik H, Sundaram R M.2012. Identification and fine-mapping of Xa33, a novel gene for resistance to Xanthomonas Oryzae pv. oryzae.Phytopathology, 102(2): 222-228. |
| 45 | Kumbhar S D, Kulwal P L, Patil J V, Sarawate C D, Gaikwad A P, Jadhav A S.2015. Genetic diversity and population structure in landraces and improved rice varieties from India.Rice Sci, 22(3): 99-107. |
| 46 | Ling K C, Aguicro V M, Lee S H.1970. A mass screening method for testing resistance to grassy stunt disease of rice.Plant Dis Rep, 54(7): 565-569. |
| 47 | Liu R, Zheng X M, Zhou L, Zhou H F, Ge S.2015. Population genetic structure of Oryza rufipogon and O. nivara: Implications for the origin of O. nivara.Mol Ecol, 24: 5211-5228. |
| 48 | Londo J P, Chiang Y C, Hung K H, Chiang T Y, Schaal B A.2006. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa.Proc Natl Acad Sci USA, 103: 9578-9583. |
| 49 | Lu B R, Song Z P, Chen J K.2002. Gene flow from crops to wild relatives in Asia: Case studies and general expectations. The 7th International Symposium on the Biosafety of Genetically Modified Organisms, Beijing, China, 10-16 October: 29-36. |
| 50 | Marathi B, Jena K K.2015. Floral traits to enhance outcrossing for higher hybrid seed production in rice: Present status and future prospects.Euphytica, 201: 1-14. |
| 51 | Marri P R, Sarla N, Reddy L V, Siddiq E A.2005. Identification and mapping of yield and yield related QTLs from an Indian accession of Oryza rufipogon.BMC Genet, 6: 33. |
| 52 | Milbourne D, Meyer R, Bradshaw J E, Baird E, Bonar N, Provan J, Powell W, Waugh R.1997. Comparison of PCR-based marker system for the analysis of genetic relationships in cultivated potato.Mol Breeding, 3(2): 127-136. |
| 53 | Nagaraju J, Kathirvel M, Kumar R R, Siddiq E A, Hasnain S E.2002. Genetic analysis of traditional and evolved basmati and non-basmati rice varieties by using fluorescence-based ISSR-PCR and SSR markers.Proc Natl Acad Sci USA, 99: 5836-5841. |
| 54 | Natrajkumar P, Sujatha K, Laha G S, Viraktamath B C, Mishra B, Srinivasa Rao K, Hari Y, Hajira S, Pranathi K, Balachiranjeevi C H, Yugander A, Sama V S A K, Balachandran S M, Sheshu Madhav M, Ram T, ShobhaRani N, Neeraja C N, Mangrauthia S K, Reddy G A, Sundaram R M.2011. Identification, molecular mapping and marker-assisted introgression of novel bacterial blight resistance genes from wild relatives of Oryza.Ind J Genet Plant Breeding, 71(2): 1-8. |
| 55 | Neelamraju S, Swamy B P M, Kaladhar K, Anuradha K, Venkateshwar Rao Y, Batchu A K, Agarwal S, Babu A P, Sudhakar T, Sreenu K, Longvah T, Surekha K, Rao K V, Reddy G A, Roja T V, Kiranmayi S L, Radhika K, Manorama K, Cheralu C, Viraktamath B C.2012. Increasing iron and zinc in rice grains using deep water rices and wild species: Identifying genomic segments and candidate genes.Qual Assur Saf Crop Foods, 4(3): 136-158. |
| 56 | Neeraja C N, Sarla N, Siddiq E A.2002. RAPD analysis of genetic diversity in Indian landraces of rice (Oryza sativa L.).J Plant Biochem Biot, 11(2): 93-97. |
| 57 | Oka H I.1988. Origin of Cultivated Rice. Tokyo, Japan: Japanese Science Society Press: 254. |
| 58 | Oka H I, Morishima H.1967. Variations in the breeding systems of wild rice, Oryza perennis.Evolution, 21(2): 249-258. |
| 59 | Panda S, Naik D, Kamble A.2015. Population structure and genetic diversity of the perennial medicinal shrub Plumbago. AoB Plants, 7: plv048. |
| 60 | Parsons B J, Newbury H J, Jackson M T, Ford-Lloyd B V.1997. Contrasting genetic diversity relationships are revealed in rice (Oryza sativa L.) using different marker types.Mol Breeding, 3(2): 115-125. |
| 61 | Perrier X, Flori A, Bonnot F.2003. Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann J C. Genetic Diversity of Cultivated Tropical Plants. Montpellier, France: Science: 43-76. |
| 62 | Phan P D T, Kageyama H, Ishikawa R, Ishii T.2012. Estimation of the outcrossing rate for annual Asian wild rice under field conditions.Breeding Sci, 62(3): 256-262. |
| 63 | Prathepha P.2012. Genetic diversity and population structure of wild rice, Oryza rufipogon from north eastern Thailand and Laos.Aust J Crop Sci, 6: 717-723. |
| 64 | Prevost A, Wilkinson M J.1999. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars.Theor Appl Genet, 98(1): 107-112. |
| 65 | Pritchard J K, Stephens M, Donnelly P.2000. Inference of population structure using multi locus genotype data.Genetics, 155(2): 945-959. |
| 66 | Qian W, Ge S, Hong D Y.2001. Genetic variation within and among populations of a wild rice Oryza granulata from China detected by RAPD and ISSR markers.Theor Appl Genet, 102(2): 440-449. |
| 67 | Rabbani M A, Pervaiz Z H, Masood M S.2008. Genetic diversity analysis of traditional and improved cultivars of Pakistani rice (Oryza sativa L.) using RAPD markers.Elect J Biotechnol, 11(3): 1-10. |
| 68 | Rahman M L, Jiang W, Chu S H, Qiao Y, Ham T H, Woo M O, Lee J, Khanam M S, Chin J H, Jeung J U, Brar D S, Jena K K, Koh H J.2009. High-resolution mapping of two rice brown planthopper resistance genes, Bph20(t) and Bph21(t), originating from Oryza minuta. Theor Appl Genet, 119(7): 1237-1246. |
| 69 | Ram T, Majumder N D, Krishnaveni D, Ansari M M.2007. Rice variety Dhanrasi, an example of improving yield potential and disease resistance by introgressing gene(s) from wild species (Oryza rufipogon).Curr Sci, 92: 987-992. |
| 70 | Ram T, Majumder N D, Mishra B.2010. Jarava: A new high yielding and pest resistant rice variety for coastal saline areas.Int Rice Res Notes, 34: 1-4. |
| 71 | Ram S G, Thiruvengadam V, Vinod K K.2007. Genetic diversity among cultivars, landraces and wild relatives of rice as revealed by microsatellite markers.J Appl Genet, 48(4): 337-345. |
| 72 | Rao V R, Hodgkin T.2002. Genetic diversity and conservation and utilization of plant genetic resources.Plant Cell Tiss Organ, 68: 1-19. |
| 73 | Rathore M, Singh R, Kumar B.2013. Weedy rice: An emerging threat to rice cultivation and options for its management.Curr Sci, 105: 1067-1072. |
| 74 | Reddy C S, Babu A P, Swamy B P M, Kaladhar K, Sarla N.2009. ISSR markers based on GA and AG repeats reveal genetic relationship among rice varieties tolerant to drought, flood, or salinity.J Zhejiang Univ Sci B, 10(2): 133-141. |
| 75 | Reddy M P, Sarla N, Siddiq E A.2002. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding.Euphytica, 128(1): 9-17. |
| 76 | Ren F, Lu B R, Li S, Huang J, Zhu Y.2003. A comparative study of genetic relationships among the AA-genome Oryza species using RAPD and SSR markers.Theor Appl Genet, 108(1): 113-120. |
| 77 | Rohlf F J. 1989. Numerical Taxonomy and Multivariate Analysis System. Version 2.02i. New York, USA: Exeter Publishing. |
| 78 | Roja V, Kiranmayi S L, Sarla N.2013. Enrichment of iron and zinc concentration in introgression lines of brown rice.Trends BioSci, 6(6): 870-875. |
| 79 | Sakai K I, Narise T.1959. Studies on the breeding behavior of wild rice.Annu Rep Natl Inst Genet Jpn, 9: 64-65. |
| 80 | Sarla N, Bobba S, Siddiq E A.2003. ISSR and SSR markers based on AG and GA repeats delineate geographically diverse Oryza nivara accessions and reveal rare alleles.Curr Sci, 84: 683-690. |
| 81 | Sarla N, Neeraja C N, Siddiq E A.2005. Use of anchored (AG)n and (GA)n primers to assess genetic diversity of Indian landraces and varieties of rice.Curr Sci, 89: 1371-1381. |
| 82 | Septiningsih E M, Prasetiyono J, Lubis E, Tai T H, Tjubaryat T, Moeljopawiro S, McCouch S R.2003. Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon.Theor Appl Genet, 107(8): 1419-1432. |
| 83 | Shakiba E, Eizenga G C.2014. Unraveling the secrets of rice wild species. In: Yan W G, Bao J S. Rice: Germplasm, Genetics and Improvement. Croatia: Intech: 1-58. |
| 84 | Song Z P, Xu X, Wang B, Chen J K, Lu B R.2003. Genetic diversity in the northernmost Oryza rufipogon populations estimated by SSR markers.Theor Appl Genet, 107(8): 1492-1499. |
| 85 | Sudhakar T, Panigrahy M, Lakshmanaik M, Babu A P, Reddy C S, Anuradha K, Swamy B P M, Sarla N.2012. Variation and correlation of phenotypic traits contributing to high yield in KMR3: Oryza rufipogon introgression lines.Int J Plant Breeding Genet, 6: 69-82. |
| 86 | Sujatha K, Natarajkumar P, Laha G S, Mishra B, Srinivasarao K, Viraktamath B C, Kirti P B, Hari Y, Balachandran S M, Rajendrakumar P, Ram T, Hajira S K, Madhav M S, Neeraja C N, Sundaram R M.2011. Inheritance of bacterial blight resistance in the rice cultivar Ajaya and high-resolution mapping of a major QTL associated with resistance.Genet Res Camb, 93: 397-408. |
| 87 | Swamy B P M, Sarla N.2008. Yield enhancing QTLs from wild species.Biotechnol Adv, 26: 106-120. |
| 88 | Swamy B P M, Kaladhar K, Ramesha M S, Viraktamath B C, Sarla N.2011. Molecular mapping of QTLs for yield and related traits in Oryza sativa cv Swarna × O. nivara (IRGC81848) backcross population.Rice Sci, 18: 178-186. |
| 89 | Swamy B P M, Kaladhar K, Shobharani N, Prasad G S V, Viraktamath B C, Reddy G A, Sarla N.2012. QTL analysis for grain quality traits in 2 BC2F2 populations derived from crosses between Oryza sativa cv Swarna and 2 accessions of O. nivara.J Hered, 103: 442-452. |
| 90 | Swamy B P M, Kaladhar K, Reddy G A, Viraktamath B C, Sarla N.2014. Mapping and introgression of QTL for yield and related traits in two backcross populations derived from Oryza sativa cv. Swarna and two accessions of O. nivara.J Genet, 93: 643-654. |
| 91 | Sweeney M, McCouch S.2007. The complex history of the domestication of rice.Ann Bot, 100(5): 951-957. |
| 92 | Tanksley S D, McCouch S R.1997. Seed banks and molecular maps: Unlocking genetic potential from the wild.Science, 277: 1063-1066. |
| 93 | Thalapati S, Batchu A K, Neelamraju S, Ramanan R.2012. Os11GSK gene from a wild rice, O. rufipogon improves yield in rice.Funct Integr Genom, 12(2): 277-289. |
| 94 | Thalapati S, Guttikonda H, Nannapaneni N D, Adari P B, Reddy C S, Swamy B P M, Batchu A K, Basava R K, Viraktamath B C, Neelamraju S.2014. Heterosis and combining ability in rice as influenced by introgressions from wild species Oryza rufipogon including qyld2.1 sub-QTL into the restorer line KMR3.Euphytica, 202(1): 81-95. |
| 95 | Thomson M J, Tai T H, McClung A M, Lai X H, Hinga M E, Lobos K B, Xu Y, Martinez C P, McCouch S R.2003. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson.Theor Appl Genet, 107(3): 479-493. |
| 96 | Tian F, Li D J, Fu Q, Zhu Z F, Fu Y C, Wang X K, Sun C Q.2006. Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (O. sativa L.) background and characterization of introgressed segments associated with yield-related traits.Theor Appl Genet, 112(3): 570-580. |
| 97 | Vaughan D A.1989. The genus Oryza L. current status of taxonomy.IRRI Res Pap Ser, 138: 1-21. |
| 98 | Wu Z H, Shi J, Xi M L, Jiang F X, Deng M W, Dayanandan S.2015. Inter simple sequence repeat data reveals high genetic diversity in wild populations of the narrowly distributed endemic Lilium regalein the Minjiang river valley of China.PLoS One, 10: e0118831. |
| 99 | Xiao J H, Li J M, Grandillo S, Ahn S N, Yuan L P, Tanksley S D, McCouch S R.1998. Identification of trait-improving quantitative trait loci alleles from a wild rice relative,Oryza rufipogon. Genetics, 150(2): 899-909. |
| 100 | Xie X B, Jin F X, Song M H, Suh J P, Hwang H G, Kim Y G, McCouch S R, Ahn S N.2008. Fine mapping a yield-enhancing QTL cluster associated with transgression variation in an Oryza sativa and O. rufipogon cross.Theor Appl Genet, 116(5): 613-622. |
| 101 | Xie X B, Song M H, Jin F X, Ahn S N, Suh J P, Hwang H G, McCouch S R.2006. Fine mapping a grain weight quantitative trait loci on rice chromosome 8 using nearly-isogenic lines derived from a cross between Oryza sativa and O. rufipogon.Theor Appl Genet, 113: 885-894. |
| 102 | Yoon D B, Kang K H, Kim H J, Ju H G, Kwon S J, Suh J P, Jeong O Y, Ahn S N.2006. Mapping quantitative trait loci for yield loci for yield components and morphological traits in an advanced backcross population between Oryza grandiglumis and the O. sativa japonica cultivar Hwaseongbyeo.Theor Appl Genet, 112: 1052-1062. |
| 103 | Yu G, Olsen K M, Schaal B A.2011. Molecular evolution of the endosperm starch synthesis pathway genes in rice (Oryza sativa L.) and its wild ancestor, O. rufipogon L.Mol Biol Evol, 28(1): 659-671. |
| 104 | Zhang P, Li J Q, Li X L, Liu X D, Zhao X J, Lu Y G.2011. Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSR markers.PLoS One, 6: e27565. |
| 105 | Zhang D L, Zhang H L, Wang M X, Sun J L, Qi Y W, Wang F M, We X H, Han L Z, Wang X K, Li Z C.2009. Genetic structure and differentiation of Oryza sativa L. in China revealed by microsatellites.Theor Appl Genet, 119: 1105-1117. |
| 106 | Zhao K, Tung C W, Eizenga G C, Wright M H, Ali M L, Price A H, Norton G J, Islam M R, Reynolds A, Mezey J, McClung A M, Bustamante C D, McCouch S R.2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa.Nat Commun, 2: 467. |
| 107 | Zheng K, Subudhi P K, Domingo J, Magpantay G, Huang N.1995. Rapid DNA isolation for marker assisted selection in rice breeding.Rice Genet Newsl, 12: 255-258. |
| 108 | Zhu Q H, Zheng X M, Luo J C, Gaut B S, Ge S.2007. Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: Severe bottleneck during domestication of rice.Mol Biol Evol, 24(3): 875-888. |
| [1] | Zhou Ying, Wan Tao, Yuan Bin, Lei Fang, Chen Meijuan, Wang Qiong, Huang Ping, Kou Shuyan, Qiu Wenxiu, Liu Li. Improving Rice Blast Resistance by Mining Broad-Spectrum Resistance Genes at Pik Locus [J]. Rice Science, 2022, 29(2): 133-142. |
| [2] | Shalini Pulipati, Suji Somasundaram, Nitika Rana, Kavitha Kumaresan, Mohamed Shafi, Peter Civáň, Gothandapani Sellamuthu, Deepa Jaganathan, Prasanna Venkatesan Ramaravi, S. Punitha, Kalaimani Raju, Shrikant S. Mantri, R. Sowdhamini, Ajay Parida, Gayatri Venkataraman. Diversity of Sodium Transporter HKT1;5 in Genus Oryza [J]. Rice Science, 2022, 29(1): 31-46. |
| [3] | Mao-Sen Liu, Shih-Hsuan Tseng, Ting-Chu Chen, Mei-Chu Chung. Visualizing Meiotic Chromosome Pairing and Segregation in Interspecific Hybrids of Rice by Genomic in situ Hybridization [J]. Rice Science, 2021, 28(1): 69-80. |
| [4] | Pathaichindachote Wanwarang, Panyawut Natjaree, Sikaewtung Kannika, Patarapuwadol Sujin, Muangprom Amorntip. Genetic Diversity and Allelic Frequency of Selected Thai and Exotic Rice Germplasm Using SSR Markers [J]. Rice Science, 2019, 26(6): 393-403. |
| [5] | Donde Ravindra, Kumar Jitendra, Gouda Gayatri, Kumar Gupta Manoj, Mukherjee Mitadru, Yasin Baksh Sk, Mahadani Pradosh, Kumar Sahoo Khirod, Behera Lambodar, Kumar Dash Sushanta. Assessment of Genetic Diversity of Drought Tolerant and Susceptible Rice Genotypes Using Microsatellite Markers [J]. Rice Science, 2019, 26(4): 239-247. |
| [6] | Mukherjee Mitadru, Padhy Barada, Srinivasan Bharathkumar, Mahadani Pradosh, Yasin Baksh Sk, Donde Ravindra, Nath Singh Onkar, Behera Lambodar, Swain Padmini, Kumar Dash Sushanta. Revealing Genetic Relationship and Prospecting of Novel Donors Among Upland Rice Genotypes Using qDTY-Linked SSR Markers [J]. Rice Science, 2018, 25(6): 308-319. |
| [7] | Kazemi Sheidollah, Reza Eshghizadeh Hamid, Zahedi Morteza. Responses of Four Rice Varieties to Elevated CO2 and Different Salinity Levels [J]. Rice Science, 2018, 25(3): 142-151. |
| [8] | Xiongsiyee Vua, Rerkasem Benjavan, Veeradittakit Jeeraporn, Saenchai Chorpet, Lordkaew Sittichai, Thebault Prom-u-thai Chanakan. Variation in Grain Quality of Upland Rice from Luang Prabang Province, Lao PDR [J]. Rice Science, 2018, 25(2): 94-102. |
| [9] | Haritha G., P. M. Swamy B., L. Naik M., Jyothi B., Divya B., Malathi S., Sarla N.. Yield Traits and Associated Marker Segregation in Elite Introgression Lines Derived from O. sativa × O. nivara [J]. Rice Science, 2018, 25(1): 19-31. |
| [10] | Rekha Talukdar Preeti, Rathi Sunayana, Pathak Khanin, Kumar Chetia Sanjay, Nath Sarma Ramendra. Population Structure and Marker-Trait Association in Indigenous Aromatic Rice [J]. Rice Science, 2017, 24(3): 145-154. |
| [11] | Haritha G., Vishnukiran T., Yugandhar P., Sarla N., Subrahmanyam D.. Introgressions from Oryza rufipogon Increase Photosynthetic Efficiency of KMR3 Rice Lines [J]. Rice Science, 2017, 24(2): 85-96. |
| [12] | Anupam Alpana, Imam Jahangir, Mohammad Quatadah Syed, Siddaiah Anantha, Prasad Das Shankar, Variar Mukund, Prasad Mandal Nimai. Genetic Diversity Analysis of Rice Germplasm in Tripura State of Northeast India Using Drought and Blast Linked Markers [J]. Rice Science, 2017, 24(1): 10-20. |
| [13] | J. S. Rama Devi S., Singh Kuldeep, Umakanth B., Vishalakshi B., Renuka P., Vijay Sudhakar K., S. Prasad M., C. Viraktamath B., Ravindra Babu V., S. Madhav M.. Development and Identification of Novel Rice Blast Resistant Sources and Their Characterization Using Molecular Markers [J]. Rice Science, 2015, 22(6): 300-308. |
| [14] | Mao-bai Li, Hui Wang, Li-ming Cao. Evaluation of Population Structure, Genetic Diversity and Origin of Northeast Asia Weedy Rice Based on Simple Sequence Repeat Markers [J]. Rice Science, 2015, 22(4): 180-188. |
| [15] | D. Kumbhar Shailesh, L. Kulwal Pawan, V. Patil Jagannath, D. Sarawate Chandrakant, P. Gaikwad Anil, S. Jadhav Ashok. Genetic Diversity and Population Structure in Landraces and Improved Rice Varieties from India [J]. Rice Science, 2015, 22(3): 99-107. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||