
Rice Science ›› 2022, Vol. 29 ›› Issue (1): 31-46.DOI: 10.1016/j.rsci.2021.12.003
• Research Paper • Previous Articles Next Articles
Shalini Pulipati1, Suji Somasundaram1, Nitika Rana2, Kavitha Kumaresan3, Mohamed Shafi4, Peter Civáň5, Gothandapani Sellamuthu1,6, Deepa Jaganathan7, Prasanna Venkatesan Ramaravi8, S. Punitha1, Kalaimani Raju1, Shrikant S. Mantri2, R. Sowdhamini4, Ajay Parida9, Gayatri Venkataraman1( )
)
Received:2021-01-23
Accepted:2021-05-17
Online:2022-01-28
Published:2022-01-01
Contact:
Gayatri Venkataraman
Shalini Pulipati, Suji Somasundaram, Nitika Rana, Kavitha Kumaresan, Mohamed Shafi, Peter Civáň, Gothandapani Sellamuthu, Deepa Jaganathan, Prasanna Venkatesan Ramaravi, S. Punitha, Kalaimani Raju, Shrikant S. Mantri, R. Sowdhamini, Ajay Parida, Gayatri Venkataraman. Diversity of Sodium Transporter HKT1;5 in Genus Oryza[J]. Rice Science, 2022, 29(1): 31-46.
Add to citation manager EndNote|Ris|BibTeX
| O. sativa (Negrão et al, | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | O. sativa (Platten et al, |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S2N | S | S | S | S | S | S | S | S | S | S | S | JQ695816: N |
| F66L | F | F | F | F | F | F | F | F | F | F | F | F |
| T67K | T | T | T | T | T | T | T | T | T | T | T | T |
| A123P | A | A | A | A | A | G | G | A | A | A | T | JQ695818: P |
| D128N | D | D | D | D | D | - | - | H | H | - | H | JQ695815: N |
| S134N | S | S | S | S | S | - | - | - | - | S | - | JQ695818: N |
| *P140A | P | A | P | P | P | P | P | P | P | P | P | JQ695815, JQ695814, JQ695812, JQ695817: A |
| P144R | P | P | P | R | R | T | T | T | T | P | L | P |
| V162L | V | V | F | V | V | V | V | V | V | I | N | V |
| *R184H | R | H | R | R | R | S | S | S | G | S | S | JQ695814, JQ695812, JQ695817: H |
| S197P | S | S | S | S | S | - | - | - | - | - | - | JQ695818 |
| L231V | L | L | L | L | L | L | L | L | L | L | L | L |
| A233V | A | A | A | A | A | V | V | I | V | V | I | JQ695818 |
| *H332D | H | D | D | D | D | D | D | D | D | D | D | JQ695814, JQ695812, JQ695817, JQ695816, JQ695818: D |
| S338P | S | S | S | P | P | A | A | S | S | S | S | S |
| V349M | V | V | V | V | V | V | V | V | V | V | V | V |
| G364S | G | G | G | S | S | A | Y | Y | G | G | G | G |
| *L395V | L | V | V | V | V | V | V | V | V | V | V | All V |
| P508S | P | P | P | S | S | P | P | P | P | P | P | P |
| L531F | L | L | L | L | L | L | L | L | L | L | L | L |
| Q429K | Q | Q | K | Q | Q | Q | Q | Q | Q | Q | Q | JQ695818, JQ695816: K |
| 1, O. sativa ssp. japonica (AA); 2, O. sativa ssp. indica (AA); 3, O. nivara (AA); 4, O. glaberrima (AA); 5, O. barthii (AA); 6, O. punctate (BB); 7, O. minuta (BBCC); 8, O. officinalis (CC); 9, O. alta (CCDD); 10, O. brachyantha (FF); 11, O. coarctata (KKLL). Blue, green or yellow colours indicate conserved amino acid residues in related Oryza species. Corresponding amino acid residues in O. sativa HKT1;5 sequences (landraces/varieties) reported by Platten et al ( | ||||||||||||
Table 1. Non-synonymous amino acid substitutions in O. sativa HKT1;5 sequences reported by Negrão et al (2013) and comparison with those from Oryza species.
| O. sativa (Negrão et al, | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | O. sativa (Platten et al, |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S2N | S | S | S | S | S | S | S | S | S | S | S | JQ695816: N |
| F66L | F | F | F | F | F | F | F | F | F | F | F | F |
| T67K | T | T | T | T | T | T | T | T | T | T | T | T |
| A123P | A | A | A | A | A | G | G | A | A | A | T | JQ695818: P |
| D128N | D | D | D | D | D | - | - | H | H | - | H | JQ695815: N |
| S134N | S | S | S | S | S | - | - | - | - | S | - | JQ695818: N |
| *P140A | P | A | P | P | P | P | P | P | P | P | P | JQ695815, JQ695814, JQ695812, JQ695817: A |
| P144R | P | P | P | R | R | T | T | T | T | P | L | P |
| V162L | V | V | F | V | V | V | V | V | V | I | N | V |
| *R184H | R | H | R | R | R | S | S | S | G | S | S | JQ695814, JQ695812, JQ695817: H |
| S197P | S | S | S | S | S | - | - | - | - | - | - | JQ695818 |
| L231V | L | L | L | L | L | L | L | L | L | L | L | L |
| A233V | A | A | A | A | A | V | V | I | V | V | I | JQ695818 |
| *H332D | H | D | D | D | D | D | D | D | D | D | D | JQ695814, JQ695812, JQ695817, JQ695816, JQ695818: D |
| S338P | S | S | S | P | P | A | A | S | S | S | S | S |
| V349M | V | V | V | V | V | V | V | V | V | V | V | V |
| G364S | G | G | G | S | S | A | Y | Y | G | G | G | G |
| *L395V | L | V | V | V | V | V | V | V | V | V | V | All V |
| P508S | P | P | P | S | S | P | P | P | P | P | P | P |
| L531F | L | L | L | L | L | L | L | L | L | L | L | L |
| Q429K | Q | Q | K | Q | Q | Q | Q | Q | Q | Q | Q | JQ695818, JQ695816: K |
| 1, O. sativa ssp. japonica (AA); 2, O. sativa ssp. indica (AA); 3, O. nivara (AA); 4, O. glaberrima (AA); 5, O. barthii (AA); 6, O. punctate (BB); 7, O. minuta (BBCC); 8, O. officinalis (CC); 9, O. alta (CCDD); 10, O. brachyantha (FF); 11, O. coarctata (KKLL). Blue, green or yellow colours indicate conserved amino acid residues in related Oryza species. Corresponding amino acid residues in O. sativa HKT1;5 sequences (landraces/varieties) reported by Platten et al ( | ||||||||||||
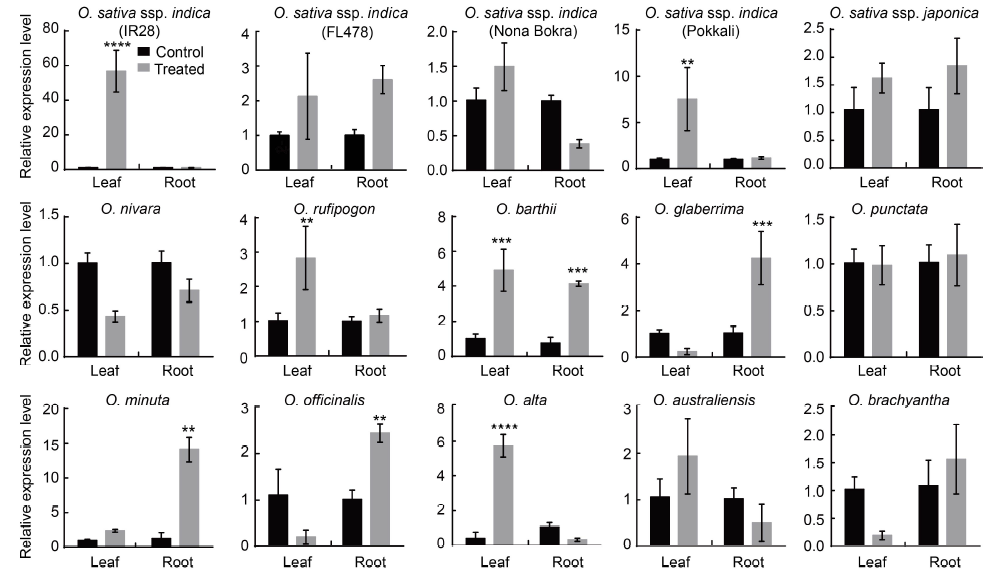
Fig. 1. HKT1;5 expression in Oryza species in leaf and root tissues under control and salinity treated conditions (incremental salinity application). Data are Mean ± SE for two biological replicates (each biological replicate was analyzed by qRT-PCR twice, each time in triplicate). Gene expression was quantified using the comparative CT (2-ΔΔCT) quantitation method with values representing ‘n’-fold difference relative to the housekeeping control gene β-Actin. Significance was calculated using the Student’s t-test. **, P < 0.01, ***, P < 0.001 and ****, P < 0.0001.
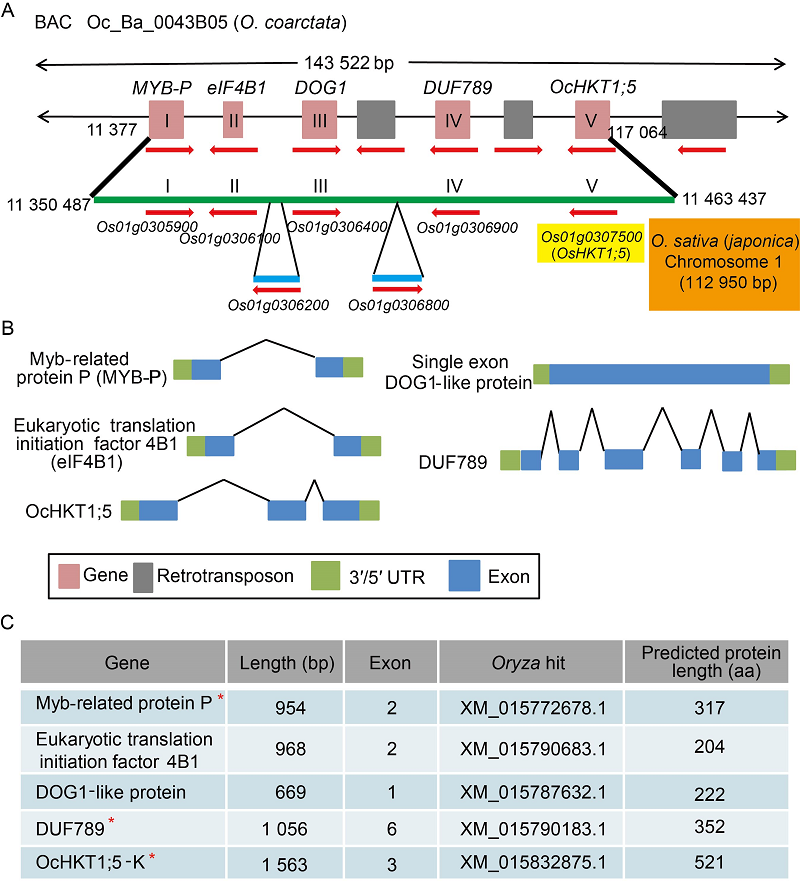
Fig. 2. O. coarctata BAC clone Oc_Ba_0043B05 sequencing and annotation. A, Genomic alignment of O. coarctata Oc_Ba_0043B05 sequence with orthologous region of O. sativa (japonica) on chromosome 1. Predicted FGNESH gene models are indicated by pink boxes while retrotransposon gene models are indicated by grey boxes. Red arrows indicate gene orientation. Orthologous region in japonica is indicated by a green line. Gene models I?V are present. In addition, two gene insertions are also present in the japonica sequence that are absent in the assembled O. coarctata BAC sequence. B, Predicted exon/intron organizations of gene models (I?V) for O. coarctata Oc_Ba_0043B05 sequence are conserved in japonica rice. C, Predicted protein lengths of models I?V and accession numbers of corresponding japonica transcript orthologs. Asterisks indicate gene models validated in O. coarctata transcriptome or by qRT-PCR (unpublished).
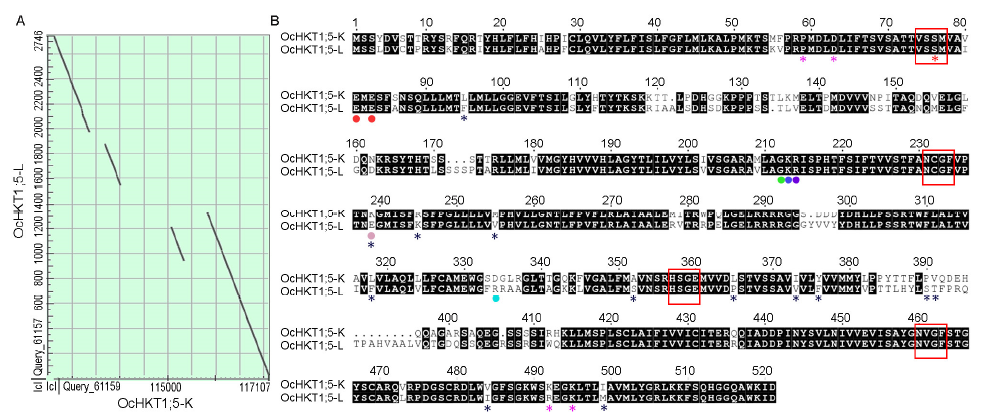
Fig. 3. Alignment of O. coarctata HKT1;5 homoeologous genomic and protein sequences. A, Dot plots showing four aligned regions of O. coarctata Oc_Ba_0043B05 sequence (143 522 bp) and OcHKT1;5-L genome (2 746 bp) sequences. BAC clone coordinates corresponding to four regions of homology (encompassing OcHKT1;5-K) with OcHKT1;5-L are indicated: (i) 115 810 to 117 107 (88% identity), (ii) 112 615 to 113 365 (87% identity), (iii) 115 067 to 115 333 (84% identity), and (iv) 113 686 to 114 009 (81% identity). B, Alignment of OcHKT1;5-K (521 amino acid residues) and OcHKT1;5-L (535 amino acid residues) translated open reading fragments. The selectivity filter residues ‘S-G-G-G’ are marked by red asterisks while the selectivity filters are boxed in red. Conserved amino acid residues near the ion pore entrance crucial to ion transport are marked by a red dot. Amino acid residues indicated by red, green, dark blue or purple dots occur near the ion pore entrance and are the same in OcHKT1;5-K and OcHKT1;5-L. Resides marked by pink and light blue dots near the ion pore entrance show opposite charges in OcHKT1;5-K and OcHKT1;5-L. Pink asterisks indicate residues that are part of an extracellular putative cation coordination site. Black asterisks indicate residues that differ between OcHKT1;5-K and OcHKT1;5-L and either occur in transmembrane segments or in loops oriented towards the ion pore entrance.
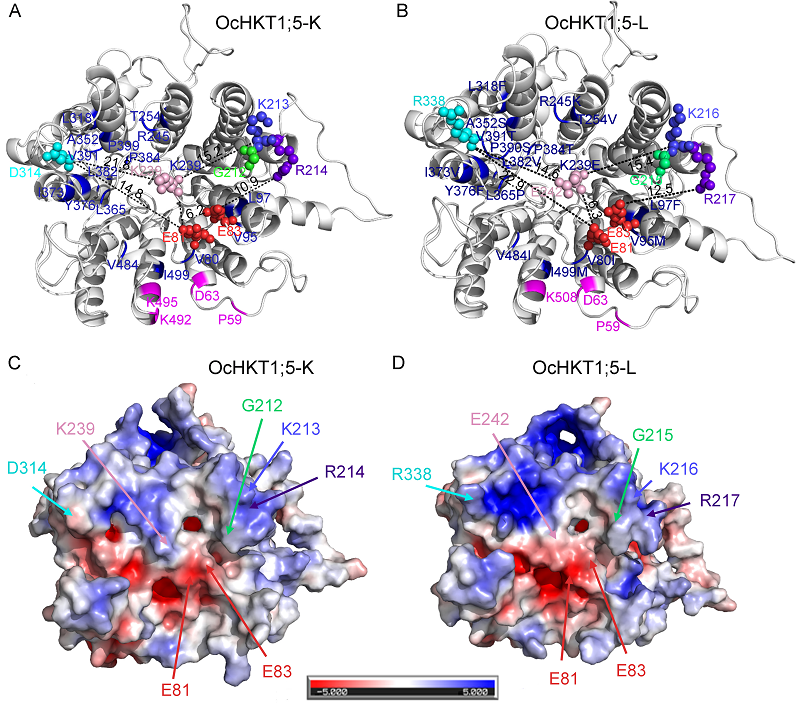
Fig. 4. Homology models and electrostatic maps of OcHKT1;5-K and OcHKT1;5-L proteins. A and B, Model monomers of OcHKT1;5-K (A) and OcHKT1;5-L (B). Key residues at the pore entrance are highlighted as spheres, and the length between residues has been measured. Extracellular cation coordination sites (magenta) and residue differences within 2 nm from the pore region of both model (blue) are highlighted. C and D, Comparative electrostatic maps of model generated for OcHKT1;5-K (C) and OcHKT1;5-L (D). Blue color indicates towards positive charge, while red color denotes negative charge. Majority of the residues at the pore region are negatively charged residues. Key amino acid residues or changes at the pore entrance have been marked.
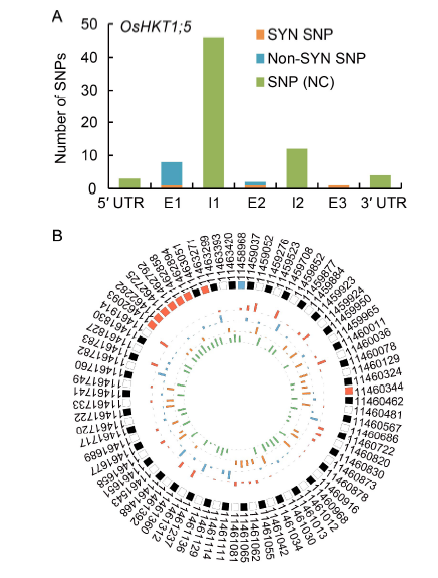
Fig. 5. SNPs identified in OsHKT1;5 in 3K-rice genomes (3K-RG). A, Distribution of SNPs in the OsHKT1;5 genomic sequence in 3K-RG. SYN SNP, Synonymous substitutions in exonic regions (E1, E2 and E3); Non-SYN SNP, Non-synonymous substitutions in exonic regions; SNP (NC), SNPs in intronic regions (I1 and I2). B, Circus based representation of SNPs and respective positions in OsHKT1;5 genomic sequence. SNP numbering as per IRGSP 1.0 (Chromosome 1). Outermost track represents SNPs and their respective positions (blue box at the top of the figure signifies the first polymorphic position; non-synonymous SNPs are indicated by orange boxes). Starting from outside, the second, third, fourth and fifth tracks represent the nucleotides adenine (red), thymine (blue), guanine (orange) and cytosine (green), respectively, with the height of each bar representing its frequency in the 3K-RG.
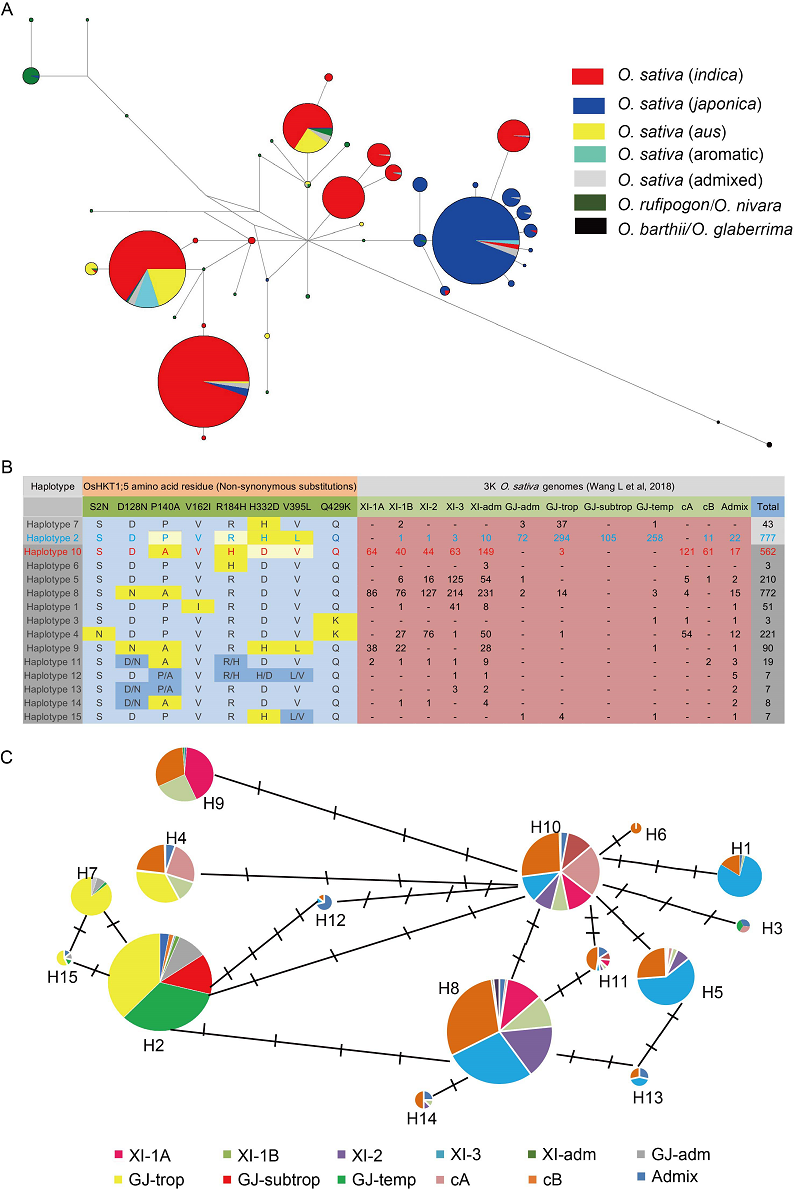
Fig. 6. Haplotyping of HKT1;5 in wild and cultivated rice. A , Only a subset of haplotypes seen in progenitor wild rice species (O. rufipogon and O. nivara) is seen in 3K-rice genomes (3K-RG). B, Haplotypes recognized with reference to 8 non-synonymous amino acid residue changes in OsHKT1;5 sequence and their distribution in 12 sub-populations (Wang W S et al, 2018) within 3K-RG [Amino acid changes indicted in bold are also referenced in Ren et al (2005)]. Haplotypes 2 (blue) and 10 (red) correspond to HKT1;5 sequences seen in salt sensitive genotype Koshihikari (also Nipponbare, IRGSP 1.0) and salt tolerant genotype Nona Bokra (Ren et al, 2005), respectively, and differ from one another in four amino acid residues indicated above. Occurrence of rare amino acid substitution in each haplotype is indicated in yellow. Haplotypes 11? 15 are heterozygous at one or more amino acid residue positions. C, Haplotype network inferred for OsHKT1;5 for the 3K-RG dataset. Each circle represents a haplotype. and the size of each circle is proportional to haplotype frequency in the population.
| [1] | Ali A, Raddatz N, Aman R, Kim S, Park H C, Jan M, Baek D, Khan I U, Oh D H, Lee S Y, Bressan R A, Lee K W, Maggio A, Pardo J M, Bohnert H J, Yun D J. 2016. A single amino-acid substitution in the sodium transporter HKT1 associated with plant salt tolerance. Plant Physiol, 171(3): 2112-2126. |
| [2] |
Almeida P M F, de Boer G J, de Boer A H. 2014. Assessment of natural variation in the first pore domain of the tomato HKT1;2 transporter and characterization of mutated versions of SlHKT1;2 expressed in Xenopus laevis oocytes and via complementation of the salt sensitive athkt1;1 mutant. Front Plant Sci, 5: 600.
PMID |
| [3] |
Ammiraju J S S, Luo M Z, Goicoechea J L, Wang W M, Kudrna D, Mueller C, Talag J, Kim H, Sisneros N B, Blackmon B, Fang E, Tomkins J B, Brar D, MacKill D, McCouch S, Kurata N, Lambert G, Galbraith D W, Arumuganathan K, Rao K, Walling J G, Gill N, Yu Y, SanMiguel P, Soderlund C, Jackson S, Wing R A. 2006. The Oryza bacterial artificial chromosome library resource: Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res, 16(1): 140-147.
PMID |
| [4] | An D, Chen J G, Gao Y Q, Li X, Chao Z F, Chen Z R, Li Q Q, Han M L, Wang Y L, Wang Y F, Chao D Y. 2017. AtHKT1 drives adaptation of Arabidopsis thaliana to salinity by reducing floral sodium content. PLoS Genet, 13(10): e1007086. |
| [5] | Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res, 44: W344-W350. |
| [6] | Asins M J, Villalta I, Aly M M, Olías R, Alvarez D E Morales P, Huertas R, Li J, Jaime-Pérez N, Haro R, Raga V, Carbonell E A, Belver A. 2013. Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+/K+ homeostasis. Plant Cell Environ, 36(6): 1171-1191. |
| [7] | Baker N, Holst M, Wang F. 2000. Adaptive multilevel finite element solution of the Poisson-Boltzmann equation: II. Refinement at solvent-accessible surfaces in biomolecular systems. J Comput Chem, 21(15): 1343-1352. |
| [8] | Bal A R, Dutt S K. 1986. Mechanism of salt tolerance in wild rice (Oryza coarctata Roxb). Plant Soil, 92: 399-404. |
| [9] |
Bandelt H J, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol, 16: 37-48.
PMID |
| [10] | Busoms S, Paajanen P, Marburger S, Bray S, Huang X Y, Poschenrieder C, Yant L, Salt D E. 2018. Fluctuating selection on migrant adaptive sodium transporter alleles in coastal Arabidopsis thaliana. Proc Natl Acad Sci USA, 115(52): E12443-E12452. |
| [11] | Byrt C S, Xu B, Krishnan M, Lightfoot D J, Athman A, Jacobs A K, Watson-Haigh N S, Plett D, Munns R, Tester M, Gilliham M. 2014. The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. Plant J, 80: 516-526. |
| [12] | Civáň P, Brown T A. 2017. Origin of rice (Oryza sativa L.) domestication genes. Genet Resour Crop Evol, 64: 1125-1132. |
| [13] | Civáň P, Craig H, Cox C J, Brown T A. 2015. Three geographically separate domestications of Asian rice. Nat Plants, 1: 15164. |
| [14] | Civáň P, Ali S, Batista-Navarro R, Drosou K, Ihejieto C, Chakraborty D, Ray A, Gladieux P, Brown T A. 2019. Origin of the aromatic group of cultivated rice (Oryza sativa L.) traced to the Indian subcontinent. Genome Biol Evol, 11(3): 832-843. |
| [15] | Cotsaftis O, Plett D, Shirley N, Tester M, Hrmova M. 2012. A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS One, 7: e39865. |
| [16] | DeLano W L. 2002. Pymol: An open-source molecular graphics tool. |
| [17] | Doyle J J, Doyle J L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull, 19: 11-15. |
| [18] | Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel R D, Bairoch A. 2003. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res, 31: 3784-3788. |
| [19] | Hoang T, Tran T, Nguyen T, Williams B, Wurm P, Bellairs S, Mundree S. 2016. Improvement of salinity stress tolerance in rice: Challenges and opportunities. Agronomy, 6(4): 54. |
| [20] |
Houston K, Qiu J E, Wege S, Hrmova M, Oakey H, Qu Y, Smith P, Situmorang A, Macaulay M, Flis P, Bayer M, Roy S, Halpin C, Russell J, Schreiber M, Byrt C, Gilliham M, Salt D E, Waugh R. 2020. Barley sodium content is regulated by natural variants of the Na+ transporter HvHKT1;5. Commun Biol, 3(1): 258.
PMID |
| [21] | Huang X H, Kurata N, Wei X H, Wang Z X, Wang A H, Zhao Q, Zhao Y, Liu K Y, Lu H Y, Li W J, Guo Y L, Lu Y Q, Zhou C C, Fan D L, Weng Q J, Zhu C R, Huang T, Zhang L, Wang Y C, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan X P, Xu Q, Dong G J, Zhan Q L, Li C Y, Fujiyama A, Toyoda A, Lu T T, Feng Q, Qian Q, Li J Y, Han B. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature, 490: 497-501. |
| [22] | Jiang Z L, Song G S, Shan X H, Wei Z Y, Liu Y Z, Jiang C, Jiang Y, Jin F X, Li Y D. 2018. Association analysis and identification of ZmHKT1;5 variation with salt-stress tolerance. Front Plant Sci, 9: 1485. |
| [23] |
Jones D T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol, 292(2): 195-202.
PMID |
| [24] | Kato N, Akai M, Zulkifli L, Matsuda N, Kato Y, Goshima S, Hazama A, Yamagami M, Guy H R, Uozumi N. 2007. Role of positively charged amino acids in the M2D transmembrane helix of Ktr/Trk/HKT type cation transporters. Channels, 1(3): 161-171. |
| [25] | Kobayashi N I, Yamaji N, Yamamoto H, Okubo K, Ueno H, Costa A, Tanoi K, Matsumura H, Fujii-Kashino M, Horiuchi T, Nayef M A, Shabala S, An G, Ma J F, Horie T. 2017. OsHKT1;5mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J, 91(4): 657-670. |
| [26] | Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol, 35: 1547-1549. |
| [27] | Kumar V, Singh A, Mithra S V A, Krishnamurthy S L, Parida S K, Jain S, Tiwari K K, Kumar P, Rao A R, Sharma S K, Khurana J P, Singh N K, Mohapatra T. 2015. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res, 22(2): 133-145. |
| [28] |
Librado P, Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25: 1451-1452.
PMID |
| [29] | Menguer P K, Sperotto R A, Ricachenevsky F K. 2017. A walk on the wild side: Oryza species as source for rice abiotic stress tolerance. Genet Mol Biol, 40: 238-252. |
| [30] | Mishra S, Singh B, Panda K, Singh B P, Singh N, Misra P, Rai V, Singh N K. 2016. Association of SNP haplotypes of HKT family genes with salt tolerance in Indian wild rice germplasm. Rice, 9: 15. |
| [31] |
Mondal T K, Rawal H C, Chowrasia S, Varshney D, Panda A K, Mazumdar A, Kaur H, Gaikwad K, Sharma T R, Singh N K. 2018. Draft genome sequence of first monocot-halophytic species Oryza coarctata reveals stress- specific genes. Sci Rep, 8: 13698.
PMID |
| [32] | Moradi F, Ismail A M, Gregorio G B, Egdane J A. 2003. Salinity tolerance of rice during reproductive development and association with tolerance at the seedling stage. Ind J Plant Physiol, 8: 105-116. |
| [33] |
Munns R, James R A, Xu B, Athman A, Conn S J, Jordans C, Byrt C S, Hare R A, Tyerman S D, Tester M, Plett D, Gilliham M. 2012. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol, 30(4): 360-364.
PMID |
| [34] |
Negrão S, Almadanim M C, Pires I S, Abreu I A, Maroco J, Courtois B, Gregorio G B, McNally K L, Oliveira M M. 2013. New allelic variants found in key rice salt-tolerance genes: An association study. Plant Biotechnol J, 11(1): 87-100.
PMID |
| [35] |
Platten J D, Cotsaftis O, Berthomieu P, Bohnert H, Davenport R J, Fairbairn D J, Horie T, Leigh R A, Lin H X, Luan S, Mäser P, Pantoja O, Rodríguez-Navarro A, Schachtman D P, Schroeder J I, Sentenac H, Uozumi N, Véry A A, Zhu J K, Dennis E S, Tester M. 2006. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci, 11(8): 372-374.
PMID |
| [36] |
Platten J D, Egdane J A, Ismail A M. 2013. Salinity tolerance, Na+ exclusion and allele mining of HKT1;5 in Oryza sativa and O. glaberrima: Many sources, many genes, one mechanism? BMC Plant Biol, 13: 32.
PMID |
| [37] |
Prusty M R, Kim S R, Vinarao R, Entila F, Egdane J, Diaz M G Q, Jena K K. 2018. Newly identified wild rice accessions conferring high salt tolerance might use a tissue tolerance mechanism in leaf. Front Plant Sci, 9: 417.
PMID |
| [38] |
Rajakani R, Sellamuthu G, Saravanakumar V, Kannappan S, Shabala L, Meinke H, Chen Z H, Zhou M X, Parida A, Shabala S, Venkataraman G. 2019. Microhair on the adaxial leaf surface of salt secreting halophytic Oryza coarctata Roxb. show distinct morphotypes: Isolation for molecular and functional analysis. Plant Sci, 285: 248-257.
PMID |
| [39] | Ren Z H, Gao J P, Li L G, Cai X L, Huang W, Chao D Y, Zhu M Z, Wang Z Y, Luan S, Lin H X. 2005. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet, 37: 1141-1146. |
| [40] |
Riedelsberger J, Vergara-Jaque A, Piñeros M, Dreyer I, González W. 2019. An extracellular cation coordination site influences ion conduction of OsHKT2;2. BMC Plant Biol, 19: 316.
PMID |
| [41] | Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee B H, Matsumoto T K, Koiwa H, Zhu J K, Bressan R A, Hasegawa P M. 2001. AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA, 98: 14150-14155. |
| [42] | Rus A, Lee B H, Muñoz-Mayor A, Sharkhuu A, Miura K, Zhu J K, Bressan R A, Hasegawa P M. 2004. AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol, 136(1): 2500-2511. |
| [43] | Šali A, Blundell T L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol, 234(3): 779-815. |
| [44] |
Sengupta S, Majumder A L. 2009. Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: A physiological and proteomic approach. Planta, 229(4): 911-929.
PMID |
| [45] | Soderlund C, Bomhoff M, Nelson W M. 2011. SyMAP v3.4: A turnkey synteny system with application to plant genomes. Nucleic Acids Res, 39(10): e68. |
| [46] | Solovyev V, Kosarev P, Seledsov I, Vorobyev D. 2006. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol, 7(Suppl 1): S10. 1-12. |
| [47] |
Somasundaram S, Véry A A, Vinekar R S, Ishikawa T, Kumari K, Pulipati S, Kumaresan K, Corratgé-Faillie C, Sowdhamini R, Parida A, Shabala L, Shabala S, Venkataraman G. 2020. Homology modeling identifies crucial amino-acid residues that confer higher Na+ transport capacity of OcHKT1;5 from Oryza coarctata Roxb. Plant Cell Physiol, 61(7): 1321-1334.
PMID |
| [48] | Stein J C, Yu Y, Copetti D, Zwickl D J, Zhang L, Zhang C J, Chougule K, Gao D Y, Iwata A, Goicoechea J L, Wei S, Wang J, Liao Y, Wang M H, Jacquemin J L, Becker C, Kudrna D, Zhang J W, Londono C E M, Song X, Lee S, Sanchez P, Zuccolo A, Ammiraju J S S, Talag J, Danowitz A, Rivera L F, Gschwend A R, Noutsos C, Wu C C, Kao S M, Zeng J W, Wei F J, Zhao Q, Feng Q, Baidouri M E, Carpentier M C, Lasserre E,Cooke R,da Rosa Farias D,da Maia L C,dos Santos R S,Nyberg K G,McNally K L,Mauleon R,Alexandrov N,Schmutz J,Flowers D,Fan C Z,Weigel D,Jena K K,Wicker T,Chen M S,Han B,Henry R,Hsing Y C,Kurata N,de Oliveira A C,Panaud O,Jackson S A,Machado C A,Sanderson M J,Long M Y,Ware D,Wing R A, 2018. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet, 50(2): 285-296. |
| [49] | Su Y, Luo W G, Lin W H, Ma L Y, Kabir M H. 2015. Model of cation transportation mediated by high-affinity potassium transporters (HKTs) in higher plants. Biol Proced Online, 17(1): 1-3. |
| [50] | The 3 000 rice genomes project. 2014. The 3 000 rice genomes project. Gigascience, 3: 7. |
| [51] | Thomson M J, de Ocampo M, Egdane J, Rahman M A, Sajise A G, Adorada D L, Tumimbang-Raiz E, Blumwald E, Seraj Z I, Singh R K, Gregorio G B, Ismail A M. 2010. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice, 3(2/3): 148-160. |
| [52] | van Bezouw R F H M, Janssen E M, Ashrafuzzaman M, Ghahramanzadeh R, Kilian B, Graner A, Visser R G F, van der Linden C G. 2019. Shoot sodium exclusion in salt stressed barley (Hordeum vulgare L.) is determined by allele specific increased expression of HKT1;5. J Plant Physiol, 241: 153029. |
| [53] | Véry A A, Nieves-Cordones M, Daly M, Khan I, Fizames C, Sentenac H. 2014. Molecular biology of K+ transport across the plant cell membrane: What do we learn from comparison between plant species? J Plant Physiol, 171(9): 748-769. |
| [54] | Wang B S, Ding Z Y, Liu W, Pan J, Li C B, Ge S, Zhang D M. 2009. Polyploid evolution in Oryza officinalis complex of the genus Oryza. BMC Evol Biol, 9(1): 250. |
| [55] |
Wang L, Liu Y H, Feng S J, Wang Z Y, Zhang J W, Zhang J L, Wang D, Gan Y T. 2018. AtHKT1 gene regulating K+ state in whole plant improves salt tolerance in transgenic tobacco plants. Sci Rep, 8: 16585.
PMID |
| [56] | Wang W S, Mauleon R, Hu Z Q, Chebotarov D, Tai S S, Wu Z C, Li M, Zheng T Q, Fuentes R R, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis K C, Xu J L, Sun C, Fu B Y, Zhang H L, Gao Y M, Zhao X Q, Shen F, Cui X, Yu H, Li Z C, Chen M L, Detras J, Zhou Y L, Zhang X Y, Zhao Y, Kudrna D, Wang C C, Li R, Jia B, Lu J Y, He X C, Dong Z T, Xu J B, Li Y H, Wang M, Shi J X, Li J, Zhang D B, Lee S, Hu W S, Poliakov A, Dubchak I, Ulat V J, Borja F N, Mendoza J R, Ali J, Li J, Gao Q, Niu Y C, Yue Z, Naredo M E B, Talag J, Wang X Q, Li J J, Fang X D, Yin Y, Glaszmann J C, Zhang J W, Li J Y, Hamilton R S, Wing R A, Ruan J, Zhang G Y, Wei C C, Alexandrov N, McNally K L, Li Z K, Leung H. 2018. Genomic variation in 3 010 diverse accessions of Asian cultivated rice. Nature, 557: 43-49. |
| [57] |
Waters S, Gilliham M, Hrmova M. 2013. Plant high-affinity potassium (HKT) transporters involved in salinity tolerance: Structural insights to probe differences in ion selectivity. Int J Mol Sci, 14(4): 7660-7680.
PMID |
| [58] | Wiederstein M, Sippl M J. 2007. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res, 35: W407-W410. |
| [59] | Williams C J, Headd J J, Moriarty N W, Prisant M G, Videau L L, Deis L N, Verma V, Keedy D A, Hintze B J, Chen V B, Jain S, Lewis S M, Arendall W B 3rd, Snoeyink J, Adams P D, Lovell S C, Richardson J S, Richardson D C. 2018. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci, 27(1): 293-315. |
| [60] | Yang C W, Zhao L, Zhang H K, Yang Z Z, Wang H, Wen S S, Zhang C Y, Rustgi S, von Wettstein D, Liu B. 2014. Evolution of physiological responses to salt stress in hexaploid wheat. Proc Natl Acad Sci USA, 111(32): 11882-11887. |
| [61] | Yang M, Lu K, Zhao F J, Xie W B, Ramakrishna P, Wang G Y, Du Q Q, Liang L M, Sun C J, Zhao H, Zhang Z Y, Liu Z H, Tian J J, Huang X Y, Wang W S, Dong H X, Hu J T, Ming L C, Xing Y Z, Wang G W, Xiao J H, Salt D E, Lian X M. 2018. Genome-wide association studies reveal the genetic basis of ionomic variation in rice. Plant Cell, 30(11): 2720-2740. |
| [1] | B. M. Lokeshkumar, S. L. Krishnamurthy, Suman Rathor, Arvinder Singh Warriach, N. M. Vinaykumar, B. M. Dushyanthakumar, Parbodh Chander Sharma. Morphophysiological Diversity and Haplotype Analysis of Saltol QTL Region in Diverse Rice Landraces for Salinity Tolerance [J]. Rice Science, 2023, 30(4): 306-320. |
| [2] | Yang Lv, Yueying Wang, Jahan Noushin, Haitao Hu, Ping Chen, Lianguang Shang, Haiyan Lin, Guojun Dong, Jiang Hu, Zhenyu Gao, Qian Qian, Yu Zhang, Longbiao Guo. Genome-Wide Association Analysis and Allelic Mining of Grain Shape-Related Traits in Rice [J]. Rice Science, 2019, 26(6): 384-392. |
| [3] | F. Aala Jr Wilson, B. Gregorio Glenn. Morphological and Molecular Characterization of Novel Salt-Tolerant Rice Germplasms from the Philippines and Bangladesh [J]. Rice Science, 2019, 26(3): 178-188. |
| [4] | Yaobin Qin, Peng Cheng, Yichen Cheng, Yue Feng, Derun Huang, Tingxu Huang, Xianjun Song, Jiezheng Ying. QTL-Seq Identified a Major QTL for Grain Length and Weight in Rice Using Near Isogenic F2 Population [J]. Rice Science, 2018, 25(3): 121-131. |
| [5] | Chandra Roy Subhas, Bhasker Reddy Lachagari Vijaya. Assessment of SNP and InDel Variations Among Rice Lines of Tulaipanji x Ranjit [J]. Rice Science, 2017, 24(6): 336-348. |
| [6] | S. M. Masuduzzaman A., Maksudul Haque Md., K. M. Shamsuddin A., A. Salam M., Ansar Ali Md.. Haplotype Diversity at Sub1 Locus and Allelic Distribution Among Rice Varieties of Tide and Flood Prone Areas of South-East Asia [J]. Rice Science, 2017, 24(4): 198-206. |
| [7] | Quynh-Hoa Pham, Xuan-An Tran, Thi-Nha-Trang Nguyen, Thi-Thuy-Anh Tran, Hai-Yen Hoang, Thi-Hong-Van Nguyen, Thi-Hanh Tang, Thi-Phuc Do. Investigation of Polymorphisms in Coding Region of OsHKT1 in Relation to Salinity in Rice [J]. Rice Science, 2016, 23(6): 334-338. |
| [8] | D. Chowdhury A., Haritha G., Sunitha T., L. Krishnamurthy S., Divya B., Padmavathi G., Ram T., Sarla N.. Haplotyping of Rice Genotypes Using Simple Sequence Repeat Markers Associated with Salt Tolerance [J]. Rice Science, 2016, 23(6): 317-325. |
| [9] | Swar Oo Kyaw, Kongjaimun Alisa, Khanthong Srisawat, Yi Myint, Tin Myint Tin, Korinsak Siriporn, Lanceras Siangliw Jonaliza, Myo Myint Khin, Vanavichit Apichart, Malumpong Chanate, Toojinda Theerayut. Characterization of Myanmar Paw San Hmwe Accessions Using Functional Genetic Markers [J]. Rice Science, 2015, 22(2): 53-64. |
| [10] | Md. Nasim ALI, Bhaswati GHOSH, Saikat GANTAIT, Somsubhra CHAKRABORTY. Selection of Rice Genotypes for Salinity Tolerance Through Morpho-Biochemical Assessment [J]. RICE SCIENCE, 2014, 21(5): 288-298. |
| [11] | XU Fei-fei #, TANG Fu-fu #, SHAO Ya-fang, CHEN Ya-ling, TONG Chuan, BAO Jin-song. Genotype X Environment Interactions for Agronomic Traits of Rice Revealed by Association Mapping [J]. RICE SCIENCE, 2014, 21(3): 133-141. |
| [12] | HE Zhi-zhou1, 2, XIE Fang-ming2, CHEN Li-yun1, Madonna Angelita DELA PAZ2. Genetic Diversity of Tropical Hybrid Rice Germplasm Measured by Molecular Markers [J]. RICE SCIENCE, 2012, 19(3): 193-201. |
| [13] | PAN Cun-hong, LI Ai-hong, DAI Zheng-yuan, ZHANG Hong-xi, LIU Guang-qing, WANG Zi-bin, MA Yu-yin, YIN Yue-jun, ZHANG Ya-fang, ZUO Shi-min, CHEN Zong-xiang, PAN Xue-biao. InDel and SNP Markers and Their Applications in Map-based Cloning of Rice Genes [J]. RICE SCIENCE, 2008, 15(4): 251-258 . |
| [14] | WAN Ying-xiu, DENG Qi-ming, WANG Shi-quan, LIU Ming-wei, ZHOU Hua-qiang, LI Ping , . Genetic Polymorphism of Wx Gene and Its Correlation with Main Grain Quality Characteristics in Rice [J]. RICE SCIENCE, 2007, 14(2): 85-93 . |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||