
Rice Science ›› 2021, Vol. 28 ›› Issue (1): 81-88.DOI: 10.1016/j.rsci.2020.11.009
• Research Papers • Previous Articles Next Articles
Shilin Ding1,2, Chaolei Liu2, Lianguang Shang3, Shenglong Yang2, Anpeng Zhang2, Hongzhen Jiang2, Banpu Ruan2, Guonan Fang2, Biao Tian2, Guoyou Ye4, Longbiao Guo2, Qian Qian2( ), Zhenyu Gao2(
), Zhenyu Gao2( )
)
Received:2019-12-12
Accepted:2020-03-30
Online:2021-01-28
Published:2021-01-28
About author:#These authors contributed equally to this work
Shilin Ding, Chaolei Liu, Lianguang Shang, Shenglong Yang, Anpeng Zhang, Hongzhen Jiang, Banpu Ruan, Guonan Fang, Biao Tian, Guoyou Ye, Longbiao Guo, Qian Qian, Zhenyu Gao. Identification of QTLs for Cadmium Tolerance During Seedling Stage and Validation of qCDSL1 in Rice[J]. Rice Science, 2021, 28(1): 81-88.
Add to citation manager EndNote|Ris|BibTeX
| Cd content (μmol/L) | Root length (cm) | Shoot length (cm) | Root dry weight (mg) | Shoot dry weight (mg) | Total dry weight (mg) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 93-11 | PA64s | 93-11 | PA64s | 93-11 | PA64s | 93-11 | PA64s | 93-11 | PA64s | |||||||||||||
| 0 | 13.34 ± 0.85 | 11.15 ± 1.09 | 27.27 ± 1.23 | 31.15 ± 1.23** | 21.8 ± 1.1 | 18.8 ± 1.2** | 91.4 ± 9.4 | 69.0 ± 5.3** | 113.2 ± 6.7 | 87.8 ± 2.3** | ||||||||||||
| 10 | 11.59 ± 0.77 | 11.35 ± 0.39 | 24.06 ± 1.12 | 27.83 ± 1.29* | 18.1 ± 1.7 | 18.4 ± 0.3 | 67.3 ± 3.2 | 62.0 ± 2.2** | 84.4 ± 2.5 | 80.4 ± 4.5* | ||||||||||||
| 20 | 8.33 ± 0.87 | 10.74 ± 0.87* | 23.86 ± 0.98 | 26.62 ± 1.98* | 16.1 ± 1.5 | 18.1 ± 0.4* | 67.0 ± 2.1 | 61.4 ± 3.5** | 83.1 ± 6.3 | 79.5 ± 1.9* | ||||||||||||
| 30 | 8.20 ± 0.74 | 9.60 ± 0.57* | 23.75 ± 0.99 | 26.59 ± 1.23* | 15.0 ± 1.0 | 17.9 ± 0.9* | 67.0 ± 2.3 | 60.2 ± 2.4** | 82.0 ± 6.5 | 78.1 ± 2.3* | ||||||||||||
| 40 | 8.10 ± 0.57 | 9.55 ± 0.91* | 23.45 ± 1.33 | 26.78 ± 1.32* | 12.0 ± 0.9 | 17.7 ± 0.2** | 66.1 ± 2.3 | 59.9 ± 3.1** | 78.1 ± 2.1 | 77.6 ± 4.4 | ||||||||||||
| 50 | 8.01 ± 0.54 | 9.52 ± 0.87* | 20.16 ± 0.56 | 26.67 ± 0.99** | 10.1 ± 0.4 | 17.6 ± 0.3** | 52.6 ± 3.7 | 59.0 ± 2.4** | 62.7 ± 3.3 | 76.6 ± 4.7** | ||||||||||||
| 100 | 7.50 ± 0.14 | 7.63 ± 0.81 | 20.93 ± 0.67 | 26.20 ± 1.12** | 7.0 ± 0.5 | 4.5 ± 0.1** | 51.4 ± 4.2 | 40.2 ± 1.2** | 58.4 ± 2.6 | 44.7 ± 3.4** | ||||||||||||
| 150 | 7.52 ± 0.69 | 7.84 ± 1.06 | 19.66 ± 0.87 | 26.15 ± 0.92** | 4.6 ± 0.3 | 3.9 ± 0.3* | 36.2 ± 5.6 | 37.1 ± 3.1 | 40.8 ± 4.1 | 41.0 ± 3.1 | ||||||||||||
Table 1 Phenotypic values of parents under different concentrations of Cd.
| Cd content (μmol/L) | Root length (cm) | Shoot length (cm) | Root dry weight (mg) | Shoot dry weight (mg) | Total dry weight (mg) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 93-11 | PA64s | 93-11 | PA64s | 93-11 | PA64s | 93-11 | PA64s | 93-11 | PA64s | |||||||||||||
| 0 | 13.34 ± 0.85 | 11.15 ± 1.09 | 27.27 ± 1.23 | 31.15 ± 1.23** | 21.8 ± 1.1 | 18.8 ± 1.2** | 91.4 ± 9.4 | 69.0 ± 5.3** | 113.2 ± 6.7 | 87.8 ± 2.3** | ||||||||||||
| 10 | 11.59 ± 0.77 | 11.35 ± 0.39 | 24.06 ± 1.12 | 27.83 ± 1.29* | 18.1 ± 1.7 | 18.4 ± 0.3 | 67.3 ± 3.2 | 62.0 ± 2.2** | 84.4 ± 2.5 | 80.4 ± 4.5* | ||||||||||||
| 20 | 8.33 ± 0.87 | 10.74 ± 0.87* | 23.86 ± 0.98 | 26.62 ± 1.98* | 16.1 ± 1.5 | 18.1 ± 0.4* | 67.0 ± 2.1 | 61.4 ± 3.5** | 83.1 ± 6.3 | 79.5 ± 1.9* | ||||||||||||
| 30 | 8.20 ± 0.74 | 9.60 ± 0.57* | 23.75 ± 0.99 | 26.59 ± 1.23* | 15.0 ± 1.0 | 17.9 ± 0.9* | 67.0 ± 2.3 | 60.2 ± 2.4** | 82.0 ± 6.5 | 78.1 ± 2.3* | ||||||||||||
| 40 | 8.10 ± 0.57 | 9.55 ± 0.91* | 23.45 ± 1.33 | 26.78 ± 1.32* | 12.0 ± 0.9 | 17.7 ± 0.2** | 66.1 ± 2.3 | 59.9 ± 3.1** | 78.1 ± 2.1 | 77.6 ± 4.4 | ||||||||||||
| 50 | 8.01 ± 0.54 | 9.52 ± 0.87* | 20.16 ± 0.56 | 26.67 ± 0.99** | 10.1 ± 0.4 | 17.6 ± 0.3** | 52.6 ± 3.7 | 59.0 ± 2.4** | 62.7 ± 3.3 | 76.6 ± 4.7** | ||||||||||||
| 100 | 7.50 ± 0.14 | 7.63 ± 0.81 | 20.93 ± 0.67 | 26.20 ± 1.12** | 7.0 ± 0.5 | 4.5 ± 0.1** | 51.4 ± 4.2 | 40.2 ± 1.2** | 58.4 ± 2.6 | 44.7 ± 3.4** | ||||||||||||
| 150 | 7.52 ± 0.69 | 7.84 ± 1.06 | 19.66 ± 0.87 | 26.15 ± 0.92** | 4.6 ± 0.3 | 3.9 ± 0.3* | 36.2 ± 5.6 | 37.1 ± 3.1 | 40.8 ± 4.1 | 41.0 ± 3.1 | ||||||||||||
| Location and treatment | Trait | Parent | RIL population | |||||
|---|---|---|---|---|---|---|---|---|
| 93-11 | PA64s | Mean | Range | Skewness | Kurtosis | |||
| Hangzhou | ||||||||
| Control | Root length (cm) | 13.70 ± 1.01 | 11.80 ± 0.51* | 11.76 | 6.35‒16.57 | -0.29 | 0.87 | |
| Shoot length (cm) | 28.83 ± 1.23 | 33.64 ± 1.31* | 31.30 | 22.31‒45.01 | 0.38 | 0.59 | ||
| Root dry weight (mg) | 13.3 ± 1.1 | 10.3 ± 1.2* | 9.1 | 2.5‒18.1 | 0.77 | 0.62 | ||
| Shoot dry weight (mg) | 44.5 ± 3.1 | 36.5 ± 2.5** | 36.3 | 10.3‒64.8 | 0.41 | 0.02 | ||
| Total dry weight (mg) | 57.8 ± 3.6 | 46.8 ± 4.1** | 45.2 | 10.3‒83.0 | 0.41 | 0.26 | ||
| Cd stress | Root length (cm) | 12.78 ± 1.30 | 11.81 ± 1.53 | 11.71 | 6.68‒17.43 | -0.03 | 0.81 | |
| Shoot length (cm) | 20.23 ± 1.01 | 24.73 ± 1.34* | 22.72 | 17.93‒29.68 | 0.52 | 0.27 | ||
| Root dry weight (mg) | 10.1 ± 1.4 | 10.6 ± 1.3 | 9.3 | 3.7‒19.2 | 0.65 | 0.40 | ||
| Shoot dry weight (mg) | 37.9 ± 2.5 | 35.4 ± 3.4 | 32.7 | 18.7‒52.8 | 0.41 | -0.38 | ||
| Total dry weight (mg) | 48.0 ± 4.1 | 46.0 ± 3.5 | 41.9 | 22.4‒71.7 | 0.36 | -0.40 | ||
| CTC | Relative root length (cm) | 0.93 ± 0.09 | 1.06 ± 0.04* | 0.98 | 0.57‒1.24 | -1.22 | 3.95 | |
| Relative shoot length (cm) | 0.70 ± 0.06 | 0.74 ± 0.09 | 0.73 | 0.55‒0.93 | -0.14 | 0.35 | ||
| Relative root dry weight (mg) | 0.76 ± 0.04 | 1.03 ± 0.07** | 1.05 | 0.26‒1.87 | 0.15 | 1.32 | ||
| Relative shoot dry weight (mg) | 0.85 ± 0.03 | 0.99 ± 0.08* | 0.90 | 0.47‒1.33 | 0.36 | 1.47 | ||
| Relative total dry weight (mg) | 0.83 ± 0.04 | 0.98 ± 0.05* | 0.93 | 0.45‒1.41 | 0.43 | 1.66 | ||
| Lingshui | ||||||||
| Control | Root length (cm) | 14.24 ± 2.24 | 11.26 ± 2.11* | 11.89 | 6.21‒17.41 | -0.26 | 1.50 | |
| Shoot length (cm) | 28.87 ± 1.23 | 31.89 ± 0.96* | 31.16 | 20.94‒44.98 | 0.42 | 0.85 | ||
| Root dry weight (mg) | 13.2 ± 0.6 | 10.4 ± 0.5* | 8.8 | 3.5‒20.5 | 0.83 | 1.77 | ||
| Shoot dry weight (mg) | 46.5 ± 3.2 | 35.7 ± 3.4** | 36.4 | 14.7‒72.4 | 0.58 | 1.03 | ||
| Total dry weight (mg) | 59.7 ± 4.0 | 46.1 ± 3.3** | 45.2 | 18.7‒92.8 | 0.65 | 1.35 | ||
| Cd stress | Root length (cm) | 13.13 ± 1.54 | 12.61 ± 2.10 | 11.80 | 6.16‒16.75 | -0.21 | 0.48 | |
| Shoot length (cm) | 18.41 ± 1.23 | 23.71 ± 1.11** | 23.07 | 15.85‒31.24 | 0.40 | 0.67 | ||
| Root dry weight (mg) | 11.4 ± 1.5 | 12.6 ± 2.2 | 9.3 | 4.5‒17.7 | 0.67 | 0.26 | ||
| Shoot dry weight (mg) | 34.4 ± 2.5 | 35.0 ± 3.3 | 32.3 | 18.1‒55.3 | 0.53 | 0.19 | ||
| Total dry weight (mg) | 45.9 ± 4.6 | 47.6 ± 4.9 | 41.6 | 23.6‒70.9 | 0.51 | -0.04 | ||
| CTC | Relative root length (cm) | 0.92 ± 0.08 | 1.12 ± 0.09** | 1.00 | 0.51‒1.20 | -1.85 | 6.10 | |
| Relative shoot length (cm) | 0.64 ± 0.06 | 0.74 ± 0.05* | 0.75 | 0.48‒0.94 | -0.26 | 0.92 | ||
| Relative root dry weight (mg) | 0.86 ± 0.09 | 1.26 ± 0.08** | 1.11 | 0.46‒1.95 | 0.70 | 1.44 | ||
| Relative shoot dry weight (mg) | 0.74 ± 0.12 | 0.98 ± 0.09** | 0.92 | 0.48‒1.48 | 0.64 | 1.60 | ||
| Relative total dry weight (mg) | 0.77 ± 0.11 | 1.00 ± 0.08** | 0.95 | 0.47‒1.45 | 0.54 | 1.00 | ||
Table 2 Phenotypic values of recombinant inbred line (RIL) population and their parents under control and Cd stress.
| Location and treatment | Trait | Parent | RIL population | |||||
|---|---|---|---|---|---|---|---|---|
| 93-11 | PA64s | Mean | Range | Skewness | Kurtosis | |||
| Hangzhou | ||||||||
| Control | Root length (cm) | 13.70 ± 1.01 | 11.80 ± 0.51* | 11.76 | 6.35‒16.57 | -0.29 | 0.87 | |
| Shoot length (cm) | 28.83 ± 1.23 | 33.64 ± 1.31* | 31.30 | 22.31‒45.01 | 0.38 | 0.59 | ||
| Root dry weight (mg) | 13.3 ± 1.1 | 10.3 ± 1.2* | 9.1 | 2.5‒18.1 | 0.77 | 0.62 | ||
| Shoot dry weight (mg) | 44.5 ± 3.1 | 36.5 ± 2.5** | 36.3 | 10.3‒64.8 | 0.41 | 0.02 | ||
| Total dry weight (mg) | 57.8 ± 3.6 | 46.8 ± 4.1** | 45.2 | 10.3‒83.0 | 0.41 | 0.26 | ||
| Cd stress | Root length (cm) | 12.78 ± 1.30 | 11.81 ± 1.53 | 11.71 | 6.68‒17.43 | -0.03 | 0.81 | |
| Shoot length (cm) | 20.23 ± 1.01 | 24.73 ± 1.34* | 22.72 | 17.93‒29.68 | 0.52 | 0.27 | ||
| Root dry weight (mg) | 10.1 ± 1.4 | 10.6 ± 1.3 | 9.3 | 3.7‒19.2 | 0.65 | 0.40 | ||
| Shoot dry weight (mg) | 37.9 ± 2.5 | 35.4 ± 3.4 | 32.7 | 18.7‒52.8 | 0.41 | -0.38 | ||
| Total dry weight (mg) | 48.0 ± 4.1 | 46.0 ± 3.5 | 41.9 | 22.4‒71.7 | 0.36 | -0.40 | ||
| CTC | Relative root length (cm) | 0.93 ± 0.09 | 1.06 ± 0.04* | 0.98 | 0.57‒1.24 | -1.22 | 3.95 | |
| Relative shoot length (cm) | 0.70 ± 0.06 | 0.74 ± 0.09 | 0.73 | 0.55‒0.93 | -0.14 | 0.35 | ||
| Relative root dry weight (mg) | 0.76 ± 0.04 | 1.03 ± 0.07** | 1.05 | 0.26‒1.87 | 0.15 | 1.32 | ||
| Relative shoot dry weight (mg) | 0.85 ± 0.03 | 0.99 ± 0.08* | 0.90 | 0.47‒1.33 | 0.36 | 1.47 | ||
| Relative total dry weight (mg) | 0.83 ± 0.04 | 0.98 ± 0.05* | 0.93 | 0.45‒1.41 | 0.43 | 1.66 | ||
| Lingshui | ||||||||
| Control | Root length (cm) | 14.24 ± 2.24 | 11.26 ± 2.11* | 11.89 | 6.21‒17.41 | -0.26 | 1.50 | |
| Shoot length (cm) | 28.87 ± 1.23 | 31.89 ± 0.96* | 31.16 | 20.94‒44.98 | 0.42 | 0.85 | ||
| Root dry weight (mg) | 13.2 ± 0.6 | 10.4 ± 0.5* | 8.8 | 3.5‒20.5 | 0.83 | 1.77 | ||
| Shoot dry weight (mg) | 46.5 ± 3.2 | 35.7 ± 3.4** | 36.4 | 14.7‒72.4 | 0.58 | 1.03 | ||
| Total dry weight (mg) | 59.7 ± 4.0 | 46.1 ± 3.3** | 45.2 | 18.7‒92.8 | 0.65 | 1.35 | ||
| Cd stress | Root length (cm) | 13.13 ± 1.54 | 12.61 ± 2.10 | 11.80 | 6.16‒16.75 | -0.21 | 0.48 | |
| Shoot length (cm) | 18.41 ± 1.23 | 23.71 ± 1.11** | 23.07 | 15.85‒31.24 | 0.40 | 0.67 | ||
| Root dry weight (mg) | 11.4 ± 1.5 | 12.6 ± 2.2 | 9.3 | 4.5‒17.7 | 0.67 | 0.26 | ||
| Shoot dry weight (mg) | 34.4 ± 2.5 | 35.0 ± 3.3 | 32.3 | 18.1‒55.3 | 0.53 | 0.19 | ||
| Total dry weight (mg) | 45.9 ± 4.6 | 47.6 ± 4.9 | 41.6 | 23.6‒70.9 | 0.51 | -0.04 | ||
| CTC | Relative root length (cm) | 0.92 ± 0.08 | 1.12 ± 0.09** | 1.00 | 0.51‒1.20 | -1.85 | 6.10 | |
| Relative shoot length (cm) | 0.64 ± 0.06 | 0.74 ± 0.05* | 0.75 | 0.48‒0.94 | -0.26 | 0.92 | ||
| Relative root dry weight (mg) | 0.86 ± 0.09 | 1.26 ± 0.08** | 1.11 | 0.46‒1.95 | 0.70 | 1.44 | ||
| Relative shoot dry weight (mg) | 0.74 ± 0.12 | 0.98 ± 0.09** | 0.92 | 0.48‒1.48 | 0.64 | 1.60 | ||
| Relative total dry weight (mg) | 0.77 ± 0.11 | 1.00 ± 0.08** | 0.95 | 0.47‒1.45 | 0.54 | 1.00 | ||
| Location and treatment | Trait | QTL a | Chr | Marker interval | Genetic distance (cM) | LOD b | Add c | Var (%) |
|---|---|---|---|---|---|---|---|---|
| Hangzhou | ||||||||
| Control | RL | qRL3 | 3 | SNP3-140‒SNP3-154 | 98.70‒113.51 | 4.58 | 1.246 | 12.6 |
| qRL4 | 4 | SNP4-90‒SNP4-127 | 29.71‒38.56 | 5.87 | -1.497 | 18.1 | ||
| qRL5 | 5 | SNP5-257‒SNP5-299 | 186.46‒202.23 | 2.89 | 0.984 | 7.8 | ||
| RDW | qRDW1.2 | 1 | SNP1-328‒SNP1-347 | 211.14‒241.11 | 3.08 | -0.002 | 11.6 | |
| SDW | qSDW2 | 2 | SNP2-252‒SNP2-269 | 136.03‒155.15 | 3.64 | -0.007 | 12.3 | |
| qSDW11.2 | 11 | SNP11-76‒SNP11-106 | 99.58‒119.73 | 3.01 | -0.006 | 10.2 | ||
| TDW | qTDW2 | 2 | SNP2-252‒SNP2-268 | 136.03‒154.58 | 3.71 | -0.009 | 12.6 | |
| qTDW11.2 | 11 | SNP11-76‒SNP11-106 | 99.58‒119.73 | 2.86 | -0.008 | 9.7 | ||
| Cd stress | RL | qCDRL2 | 2 | SNP2-265‒SNP2-288 | 150.74‒167.38 | 3.02 | -1.259 | 11.4 |
| SL | qCDSL1.1 | 1 | SNP1-311‒SNP1-347 | 201.79‒241.11 | 2.94 | -1.450 | 9.9 | |
| qCDSL5 | 5 | SNP5-275‒SNP5-299 | 194.83‒202.23 | 3.07 | 1.503 | 10.7 | ||
| RDW | qCDRDW4.1 | 4 | SNP4-1‒SNP4-29 | 0.00‒11.51 | 2.06 | -0.017 | 0. 9 | |
| qCDRDW5 | 5 | SNP5-210‒SNP5-232 | 155.42‒176.84 | 2.60 | 0.002 | 11.7 | ||
| SDW | qCDSDW2 | 2 | SNP2-252‒SNP2-267 | 136.03‒154.38 | 3.71 | -0.006 | 12.8 | |
| qCDSDW11 | 11 | SNP11-70‒SNP11-114 | 97.21‒124.18 | 2.24 | -0.005 | 7.9 | ||
| TDW | qCDTDW2 | 2 | SNP2-252‒SNP2-269 | 136.03‒155.15 | 2.58 | -0.007 | 9.2 | |
| qCDTDW11 | 11 | SNP11-70‒SNP11-114 | 97.21‒124.18 | 2.03 | -0.006 | 6.6 | ||
| CTC | RL | qRRL9 | 9 | SNP9-136‒SNP9-168 | 76.32‒97.61 | 2.26 | -0.088 | 8.8 |
| SL | qRSL9 | 9 | SNP9-160‒SNP9-180 | 90.45‒104.07 | 2.57 | 0.043 | 10.8 | |
| Lingshui Control | ||||||||
| SL | qSL3 | 3 | SNP3-110‒SNP3-121 | 73.36‒82.84 | 3.07 | 2.413 | 9.2 | |
| qSL9 | 9 | SNP9-26‒SNP9-52 | 11.10‒27.03 | 4.43 | 2.914 | 13.4 | ||
| qSL10 | 10 | SNP10-7‒SNP10-26 | 2.06‒20.03 | 3.03 | 2.374 | 8.9 | ||
| RDW | qRDW1.1 | 1 | SNP1-163‒SNP1-193 | 104.79‒127.33 | 3.78 | -0.002 | 12.5 | |
| qRDW5 | 5 | SNP5-267‒SNP5-299 | 190.72‒202.23 | 3.17 | 0.002 | 10.8 | ||
| SDW | qSDW11.1 | 11 | SNP11-39‒SNP11-57 | 32.54‒53.91 | 2.93 | 0.006 | 11.1 | |
| TDW | qTDW11.1 | 11 | SNP11-39‒SNP11-59 | 32.54‒57.47 | 2.71 | 0.008 | 10.4 | |
| Cd stress | RL | qCDRL10 | 10 | SNP10-112‒SNP10-147 | 66.22‒81.82 | 3.01 | 1.288 | 11.8 |
| SL | qCDSL1.2 | 1 | SNP1-312‒SNP1-347 | 202.18‒241.11 | 3.44 | -2.489 | 11.8 | |
| qCDSL4 | 4 | SNP4-220‒SNP4-259 | 81.03‒104.36 | 2.36 | -2.080 | 8.3 | ||
| RDW | qCDRDW4.2 | 4 | SNP4-270‒SNP4-285 | 108.33‒139.91 | 2.75 | -0.002 | 11.8 | |
| SDW | qCDSDW4 | 4 | SNP4-1‒SNP4-19 | 0.00‒8.33 | 2.67 | -0.005 | 10.5 | |
| TDW | qCDTDW4 | 4 | SNP4-1‒SNP4-19 | 0.00‒8.33 | 2.65 | -0.006 | 10.5 | |
| CTC | RL | qRRL7 | 7 | SNP7-230‒SNP7-263 | 102.30‒119.81 | 2.44 | 0.090 | 9.4 |
| RDW | qRRDW3 | 3 | SNP3-265‒SNP3-284 | 199.55‒221.41 | 3.99 | -0.241 | 15.4 | |
| SDW | qRSDW1 | 1 | SNP1-177‒SNP1-197 | 108.96‒129.65 | 2.81 | 0.141 | 10.6 | |
| TDW | qRTDW1 | 1 | SNP1-177‒SNP1-197 | 108.96‒129.65 | 2.99 | 0.149 | 11.3 |
Table 3 Putative QTLs with LOD > 2.0 detected in rice recombinant inbred line population.
| Location and treatment | Trait | QTL a | Chr | Marker interval | Genetic distance (cM) | LOD b | Add c | Var (%) |
|---|---|---|---|---|---|---|---|---|
| Hangzhou | ||||||||
| Control | RL | qRL3 | 3 | SNP3-140‒SNP3-154 | 98.70‒113.51 | 4.58 | 1.246 | 12.6 |
| qRL4 | 4 | SNP4-90‒SNP4-127 | 29.71‒38.56 | 5.87 | -1.497 | 18.1 | ||
| qRL5 | 5 | SNP5-257‒SNP5-299 | 186.46‒202.23 | 2.89 | 0.984 | 7.8 | ||
| RDW | qRDW1.2 | 1 | SNP1-328‒SNP1-347 | 211.14‒241.11 | 3.08 | -0.002 | 11.6 | |
| SDW | qSDW2 | 2 | SNP2-252‒SNP2-269 | 136.03‒155.15 | 3.64 | -0.007 | 12.3 | |
| qSDW11.2 | 11 | SNP11-76‒SNP11-106 | 99.58‒119.73 | 3.01 | -0.006 | 10.2 | ||
| TDW | qTDW2 | 2 | SNP2-252‒SNP2-268 | 136.03‒154.58 | 3.71 | -0.009 | 12.6 | |
| qTDW11.2 | 11 | SNP11-76‒SNP11-106 | 99.58‒119.73 | 2.86 | -0.008 | 9.7 | ||
| Cd stress | RL | qCDRL2 | 2 | SNP2-265‒SNP2-288 | 150.74‒167.38 | 3.02 | -1.259 | 11.4 |
| SL | qCDSL1.1 | 1 | SNP1-311‒SNP1-347 | 201.79‒241.11 | 2.94 | -1.450 | 9.9 | |
| qCDSL5 | 5 | SNP5-275‒SNP5-299 | 194.83‒202.23 | 3.07 | 1.503 | 10.7 | ||
| RDW | qCDRDW4.1 | 4 | SNP4-1‒SNP4-29 | 0.00‒11.51 | 2.06 | -0.017 | 0. 9 | |
| qCDRDW5 | 5 | SNP5-210‒SNP5-232 | 155.42‒176.84 | 2.60 | 0.002 | 11.7 | ||
| SDW | qCDSDW2 | 2 | SNP2-252‒SNP2-267 | 136.03‒154.38 | 3.71 | -0.006 | 12.8 | |
| qCDSDW11 | 11 | SNP11-70‒SNP11-114 | 97.21‒124.18 | 2.24 | -0.005 | 7.9 | ||
| TDW | qCDTDW2 | 2 | SNP2-252‒SNP2-269 | 136.03‒155.15 | 2.58 | -0.007 | 9.2 | |
| qCDTDW11 | 11 | SNP11-70‒SNP11-114 | 97.21‒124.18 | 2.03 | -0.006 | 6.6 | ||
| CTC | RL | qRRL9 | 9 | SNP9-136‒SNP9-168 | 76.32‒97.61 | 2.26 | -0.088 | 8.8 |
| SL | qRSL9 | 9 | SNP9-160‒SNP9-180 | 90.45‒104.07 | 2.57 | 0.043 | 10.8 | |
| Lingshui Control | ||||||||
| SL | qSL3 | 3 | SNP3-110‒SNP3-121 | 73.36‒82.84 | 3.07 | 2.413 | 9.2 | |
| qSL9 | 9 | SNP9-26‒SNP9-52 | 11.10‒27.03 | 4.43 | 2.914 | 13.4 | ||
| qSL10 | 10 | SNP10-7‒SNP10-26 | 2.06‒20.03 | 3.03 | 2.374 | 8.9 | ||
| RDW | qRDW1.1 | 1 | SNP1-163‒SNP1-193 | 104.79‒127.33 | 3.78 | -0.002 | 12.5 | |
| qRDW5 | 5 | SNP5-267‒SNP5-299 | 190.72‒202.23 | 3.17 | 0.002 | 10.8 | ||
| SDW | qSDW11.1 | 11 | SNP11-39‒SNP11-57 | 32.54‒53.91 | 2.93 | 0.006 | 11.1 | |
| TDW | qTDW11.1 | 11 | SNP11-39‒SNP11-59 | 32.54‒57.47 | 2.71 | 0.008 | 10.4 | |
| Cd stress | RL | qCDRL10 | 10 | SNP10-112‒SNP10-147 | 66.22‒81.82 | 3.01 | 1.288 | 11.8 |
| SL | qCDSL1.2 | 1 | SNP1-312‒SNP1-347 | 202.18‒241.11 | 3.44 | -2.489 | 11.8 | |
| qCDSL4 | 4 | SNP4-220‒SNP4-259 | 81.03‒104.36 | 2.36 | -2.080 | 8.3 | ||
| RDW | qCDRDW4.2 | 4 | SNP4-270‒SNP4-285 | 108.33‒139.91 | 2.75 | -0.002 | 11.8 | |
| SDW | qCDSDW4 | 4 | SNP4-1‒SNP4-19 | 0.00‒8.33 | 2.67 | -0.005 | 10.5 | |
| TDW | qCDTDW4 | 4 | SNP4-1‒SNP4-19 | 0.00‒8.33 | 2.65 | -0.006 | 10.5 | |
| CTC | RL | qRRL7 | 7 | SNP7-230‒SNP7-263 | 102.30‒119.81 | 2.44 | 0.090 | 9.4 |
| RDW | qRRDW3 | 3 | SNP3-265‒SNP3-284 | 199.55‒221.41 | 3.99 | -0.241 | 15.4 | |
| SDW | qRSDW1 | 1 | SNP1-177‒SNP1-197 | 108.96‒129.65 | 2.81 | 0.141 | 10.6 | |
| TDW | qRTDW1 | 1 | SNP1-177‒SNP1-197 | 108.96‒129.65 | 2.99 | 0.149 | 11.3 |
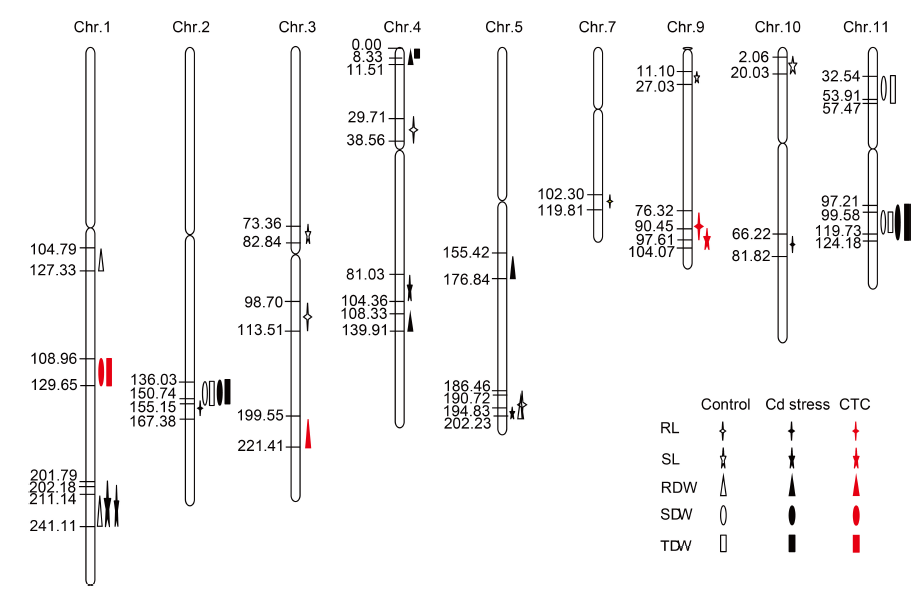
Fig. 1. Chromosomal locations of all QTLs for Cd tolerance in rice recombinant inbred line population at seedling stage.The genetic distance of marker (cM) is annotated on the left of each chromosome. Chr, Chromosome; CTC, Cd tolerance coefficient; RL, Root length; SL, Shoot length; RDW, Root dry weight; SDW, Shoot dry weight; TDW, Total dry weight.
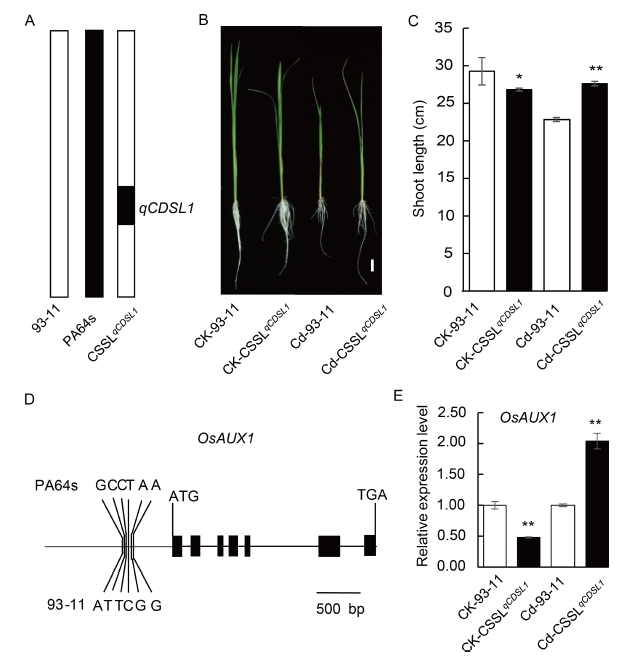
Fig. 2. Validation of qCDSL1.A, Schematic graph of chromosome 1 of CSSLqCDSL1, the parents 93-11 and PA64s. The white and black bars represent 93-11 and PA64s alleles, respectively.B, Comparison of seedling growth morphology of 93-11 and CSSLqCDSL1 under the control and Cd stress conditions. Scale bar, 2 cm.C, Shoot lengths of 93-11 and CSSLqCDSL1 under the control and Cd stress conditions. Data are Mean ± SD (n = 6). * and ** indicate 5% and 1% significant levels compared to 93-11 under the control and Cd stress conditions, respectively, according to the Student’s t-test.D, Gene structure and sequence differences of LOC_Os01g63770 (OsAUX1) between PA64s and 93-11.E, Relative expression level of OsAUX1 in shoots of 93-11 and CSSLqCDSL1 under the control and Cd stress conditions. Data are Mean ± SD (n = 3). ** indicates 1% significant level compared to 93-11 under the control and Cd stress conditions, respectively, according to the Student’s t-test.
| [1] | Agrawal G K, Rakwal R, Iwahashi H. 2002. Isolation of novel rice (Oryza sativa L.) multiple stress responsive MAP kinase gene, OsMSRMK2, whose mRNA accumulates rapidly in response to environmental cues. Biochem Biophy Res Commun, 294(5): 1009-1016. |
| [2] | Agrawal G K, Agrawal S K, Shibato J, Iwahashi H, Rakwal R, 2003. Novel rice MAP kinases OsMSRMK3 and OsWJUMK1 involved in encountering diverse environmental stresses and developmental regulation. Biochem Biophy Res Commun, 300(3): 775-783. |
| [3] | Clemens S, Aarts M G M, Thomine S, Verbruggen N. 2013. Plant science: The key to preventing slow cadmium poisoning. Trends Plant Sci, 18(2): 92-99. |
| [4] | Das N, Bhattacharya S, Bhattacharyya S, Maiti M K. 2017. Identification of alternatively spliced transcripts of rice phytochelatin synthase 2 gene OsPCS2, involved in mitigation of cadmium and arsenic stresses. Plant Mol Biol, 94: 167-183. |
| [5] | Gao Z Y, Zhao S C, He W M, Guo L B, Peng Y L, Wang J J, Guo X S, Zhang X M, Rao Y C, Zhang C, Dong G J, Zhang F Y, Lu C X, Hu J, Zhou Q, Liu H J, Wu H Y, Xu J, Ni P X, Zeng D L, Liu D H, Tian P, Gong L H, Ye C, Zhang G H, Wang J, Tian F K, Xue D W, Liao Y, Zhu L, Cheng M S, Li J Y, Cheng S H, Zhang G Y, Wang J, Qian Q. 2013. Dissecting yield-associated loci in super hybrid rice by resequencing recombinant inbred lines and improving parental genome sequences. Proc Natl Acad Sci USA, 110: 14492-14497. |
| [6] | Ishikawa S, Ishimaru Y, Igura M, Kuramata M, Abe T, Senoura T, Hase Y, Arao T, Nishizawa N K, Nakanishi H. 2012. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc Natl Acad Sci USA, 109: 19166-19171. |
| [7] | Ishimaru Y, Kakei Y, Shimo H, Bashir K, Sato Y, Sato Y, Uozumi N, Nakanishi H, Nishizawa N K. 2011. A rice phenolic efux transporter is essential for solubilizing precipitated apoplasmic iron in the plant stele. J Biol Chem, 286: 24649-24655. |
| [8] | Jarup L, Akesson A. 2009. Current status of cadmium as an environ- mental health problem. Toxicol Appl Pharmacol, 238(3): 201-208. |
| [9] | Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa N K. 2004. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J, 39(3): 415-424. |
| [10] | Kuramata M, Masuya S, Takahashi Y, Kitagawa E, Inoue C, Ishikawa S, Youssefan S, Kusano T. 2009. Novel cysteine-rich peptides from Digitaria ciliaris and Oryza sativa enhance tolerance to cadmium by limiting its cellular accumulation. Plant Cell Physiol, 50(1): 106-117. |
| [11] | Lee S, Kim Y Y, Lee Y, An G. 2007. Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol, 145(3): 831-842. |
| [12] | Li W X, Ou Yang L J, Wen W, Xiong Y Y, Xu W Q, Peng X S, Chen X R, He X P, Fu J R, Bian J M, Xu J, Zhou D H, He H H, Sun X T, Zhu C L. 2019. Identification of QTL for cadmium tolerance at seedling stage of rice (Oryza sativa L.). Acta Agric Univ Jiangxi, 41(1): 19-24. (in Chinese with English abstract) |
| [13] | Lim S D, Hwang J G, Han A R, Park Y C, Lee C, Ok Y S, Jang C S. 2014. Positive regulation of rice RING E3 ligase OsHIR1 in arsenic and cadmium uptakes. Plant Mol Biol, 85: 365-379. |
| [14] | Luo J S, Huang J, Zeng D L, Peng J S, Zhang G B, Ma H L, Guan Y, Yi H Y, Fu Y L, Han B, Lin H X, Qian Q, Gong J M. 2018. A defensin-like protein drives cadmium efflux and allocation in rice. Nat Commun, 9(1): 645. |
| [15] | McCouch S R, Cho Y G, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T. 1997. Rice: Report on QTL nomenclature. Gram Newsl, 14: 11-13. |
| [16] | Miyadate H, Adachi S, Hiraizumi A, Tezuka K, Nakazawa N, Kawamoto T, Katou K, Kodama I, Sakurai K, Takahashi H, Satoh-Nagasawa N, Watanabe A, Fujimura T, Akagi H. 2011. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol, 189(1): 190-199. |
| [17] | Mukhopadhyay A, Vij S, Tyagi A K. 2004. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA, 101(16): 6309-6314. |
| [18] | Nakanishi H, Ogawa I, Ishimaru Y, Mori S, Nishizawa N K. 2006. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe transporters OsIRT1 and OSIRT2 in rice. Soil Sci Plant Nutr, 52(4): 464-469. |
| [19] | Nawrot T S, Staessen J A, Roels H A, Munters E, Cuypers A, Richart T, Ruttens A, Smeets K, Clijsters H, Vangronsveld J. 2010. Cadmium exposure in the population: From health risks to strategies of prevention. Biometals, 23(5): 769-782. |
| [20] | Oda K, Otani M, Uraguchi S, Akihiro T, Fujiwara T. 2011. Rice ABCG43 is Cd inducible and confers Cd tolerance on yeast. Biosci Biotechnol Biochem, 75(6): 1211-1213. |
| [21] | Ramegowda Y, Venkategowda R, Jagadish P, Govind G, Hanuman- thareddy R R, Makarla U, Guligowda S. 2013. Expression of a rice Zn transporter, OsZIP1, increases Zn concentration in tobacco and finger millet transgenic plants. Plant Biotechnol Rep, 7(3): 309-319. |
| [22] | Shim D, Hwang J U, Lee J, Lee S, Choi Y, An G, Martinoia E, Lee Y. 2009. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell, 21(12): 4031-4043. |
| [23] | Shimo H, Ishimaru Y, An G, Yamakawa T, Nakanishi H, Nishizawa N K. 2011. Low cadmium (LCD), a novel gene related to cadmium tolerance and accumulation in rice. J Exp Bot, 62(15): 5727-5734. |
| [24] | Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa N K. 2011. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J Exp Bot, 62(14): 4843-4850. |
| [25] | Takahashi R, Ishimaru Y, Shimo H, Ogo Y, Senoura T, Nishizawa N K, Nakanishi H. 2012. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ, 35(11): 1948-1957. |
| [26] | Tan M P, Cheng D, Yang Y N, Zhang G Q, Qin M J, Chen J, Chen Y H, Jiang M Y. 2017. Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium responsive genes. BMC Plant Biol, 17(1): 194. |
| [27] | Uraguchi S, Kamiya T, Sakamoto T, Kassai K, Sato Y, NagamuraY, Yoshida A, Kyozuka J, Ishikawa S, Fujiwara T. 2011. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc Natl Acad Sci USA, 108: 20959-20964. |
| [28] | Wang C H, Guo W L, Shan Y, Wei P C, David W O. 2016. Reduction of Cd in rice through expression of OXS3-like gene fragments. Mol Plant, 9(2): 301-304. |
| [29] | Wang F J, Wang M, Liu Z P, Shi Y, Han T Q, Ye Y Y, Gong N, Sun J W, Zhu C. 2015. Different responses of low grain-Cd- accumulating and high grain-Cd-accumulating rice cultivars to Cd stress. Plant Physiol Biochem, 96: 261-269. |
| [30] | Xue D W, Chen M C, Zhang G P. 2009. Mapping of QTLs associated with cadmium tolerance and accumulation during seedling stage in rice (Oryza sativa L.). Euphytica, 165(3): 587-596. |
| [31] | Yan H L, Xu W X, Xie J Y, Gao Y W, Wu L L, Sun L, Feng L, Chen X, Zhang T, Dai C H, Li T, Lin X N, Zhang Z Y, Wang X Q, Li F M, Zhu X Y, Li J J, Li Z C, Chen C Y, Ma M, Zhang H L, He Z Y. 2019. Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies. Nat Commun, 10: 2562. |
| [32] | Yang M, Chen L, Xu Q G, Sun Y L. 2017. Effects of cadmium stress on seed germination and growth characteristic of different rice cultivars. Crop Res, 31(6): 659-663. |
| [33] | Yao H Y, Xu J M, Huang C Y. 2003. Substrate utilization pattern, biomass and activity of microbial communities in a sequence of heavy metal-polluted paddy soils. Geoderma, 115(1/2): 139-148. |
| [34] | Yu C L, Sun C D, Shen C J, Wang S K, Liu F, Liu Y, Chen Y L, Li C Y, Qian Q, Aryal B, Geisler M, Jiang D A, Qi Y H. 2015. The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryza sativa L.). Plant J, 83(5): 818-830. |
| [35] | Yuan L Y, Yang S G, Liu B X, Zhang M, Wu K Q. 2012. Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep, 31(1): 67-79. |
| [36] | Zhang B, Shang L G, Ruan B P, Zhang A P, Yang S L, Jiang H Z, Liu C L, Hong K, Lin H, Gao Z Y, Hu J, Zeng D L, Guo L B, Qian Q. 2019. Development of three sets of high throughput genotyped rice chromosome segment substitution lines and QTL mapping for eleven traits. Rice, 12(1): 12-33. |
| [1] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [2] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [3] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [4] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [5] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [6] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [7] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [8] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [9] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [10] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [13] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [14] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| [15] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||