
Rice Science ›› 2021, Vol. 28 ›› Issue (3): 257-267.DOI: 10.1016/j.rsci.2021.04.005
• Research Paper • Previous Articles Next Articles
Baoxiang Wang1,#, Yan Liu1,#, Yifeng Wang2,#, Jingfang Li1, Zhiguang Sun1, Ming Chi1, Yungao Xing1, Bo Xu1, Bo Yang1, Jian Li1, Jinbo Liu1, Tingmu Chen1, Zhaowei Fang1, Baiguan Lu1, Dayong Xu1( ), Kazeem Bello Babatunde1(
), Kazeem Bello Babatunde1( )
)
Received:2020-07-13
Accepted:2020-09-30
Online:2021-05-28
Published:2021-05-28
About author:#These authors contributed equally to this work
Baoxiang Wang, Yan Liu, Yifeng Wang, Jingfang Li, Zhiguang Sun, Ming Chi, Yungao Xing, Bo Xu, Bo Yang, Jian Li, Jinbo Liu, Tingmu Chen, Zhaowei Fang, Baiguan Lu, Dayong Xu, Kazeem Bello Babatunde. OsbZIP72 Is Involved in Transcriptional Gene-Regulation Pathway of Abscisic Acid Signal Transduction by Activating Rice High-Affinity Potassium Transporter OsHKT1;1[J]. Rice Science, 2021, 28(3): 257-267.
Add to citation manager EndNote|Ris|BibTeX
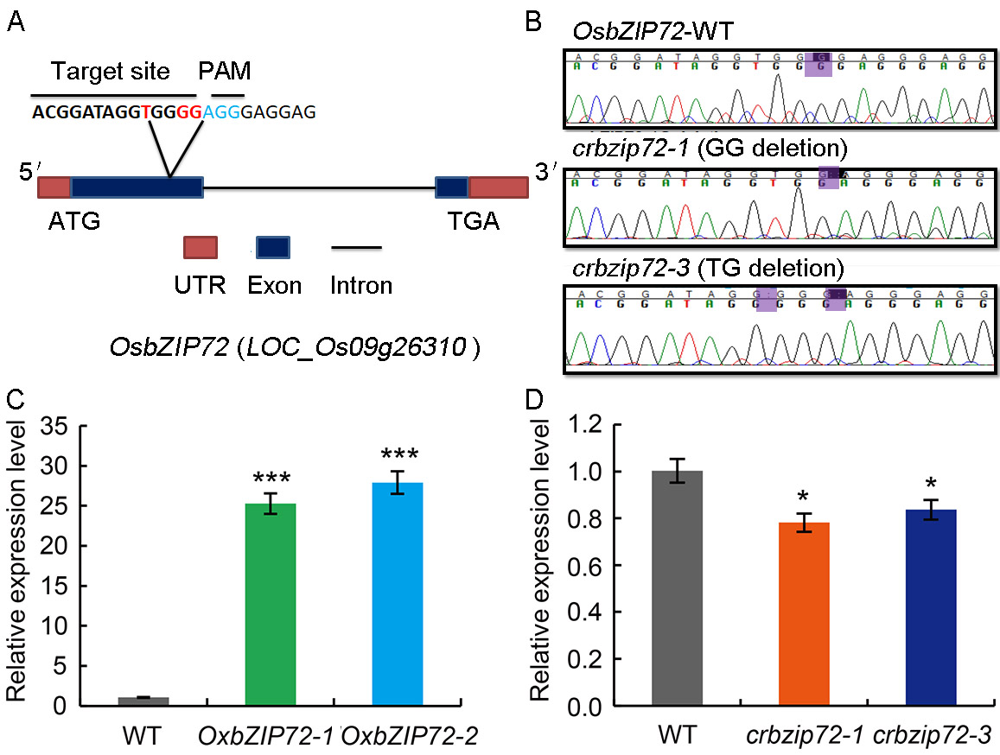
Fig. 1. Expression analysis and molecular characterization of OsbZIP72 gene. A, Schematic presentation of the OsbZIP72 gene structure and CRISPR-Cas9 editing sites. PAM, Protospacer adjacent motif; UTR, Untranslated region. B, Sanger sequencing chromatograph of the CRISPR- Cas9 target site in homozygous mutants of crbzip72. Purple letters represent the mutant sites. WT, Wild type.C, qRT-PCR analysis of OsbZIP72 gene in transgenic rice plants overexpressing OsbZIP72 (OxbZIP72-1 and OxbZIP72-2). OsActin1 was used as a reference gene. Values are Mean ± SE (n = 3). ***, P < 0.001.D, qRT-PCR analysis of OsbZIP72 in crbzip72 mutant plants (crbzip72-1 and crbzip72-3). OsActin1 was used as a reference gene. Values are Mean ± SE (n = 3). *, P < 0.05.
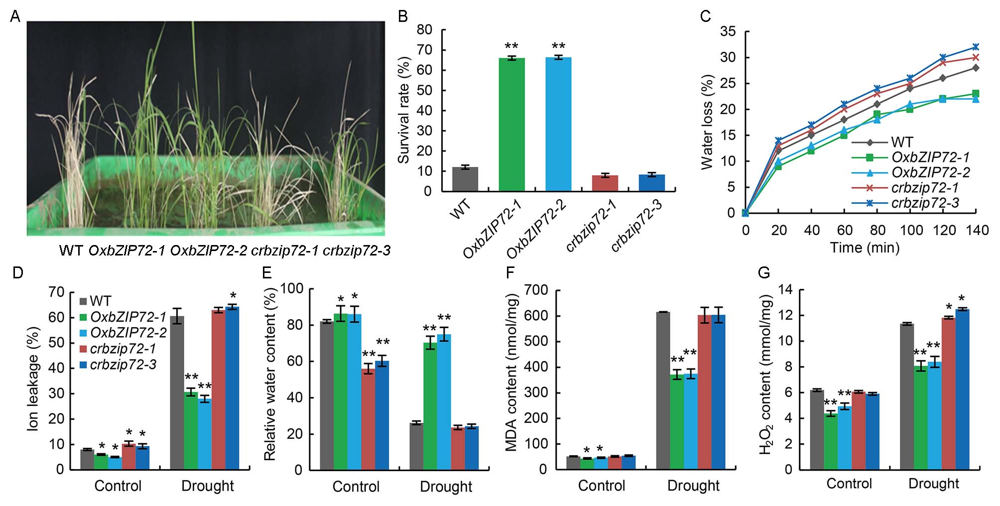
Fig. 2. Evaluation of drought and oxidative stress tolerance in OsbZIP72 overexpression (OxbZIP72) and knock-out (crbzip72) transgenic plants.A, Phenotype after withholding water for 10 d and evidence of wilting was observed in the wild type (WT).B, Survival rates of WT, OxbZIP72 and crbzip72 plants after drought stress.C, Water loss from detached leaves when WT displayed wilting after withholding irrigation. D?G, Ion leakage (D), relative water content (E), total malondialdehyde (MDA) content (F) and total H2O2 content (G) in WT, OxbZIP72 and crbzip72 plants after drought stress.Data present Mean ± SE (n = 3). ** and * indicate P < 0.01 and P < 0.05, respectively as determined by the Student’s t-test.
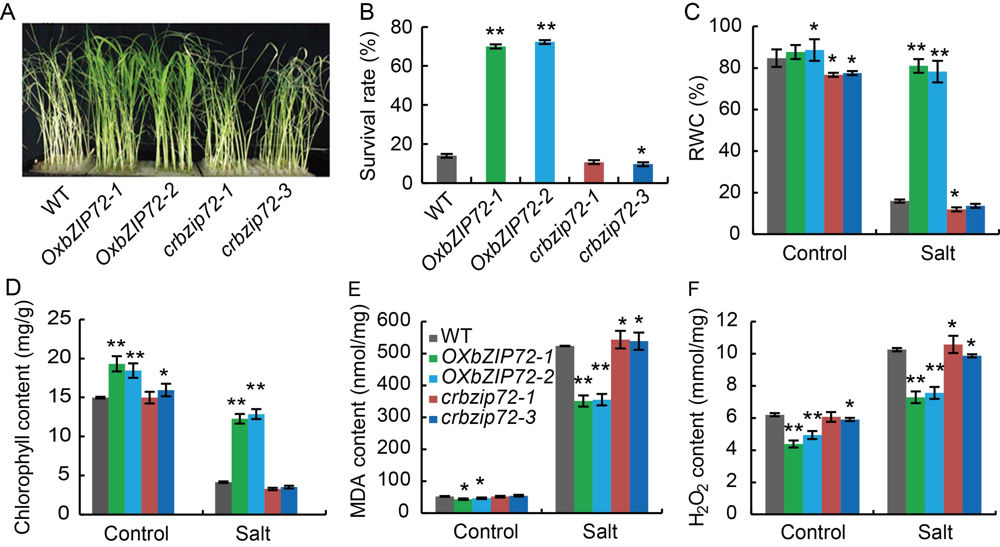
Fig. 3. Assessment of salinity stress tolerance in OsbZIP72 overexpression (OxbZIP72) and knock-out (crbzip72) transgenic plants. A, Wild type (WT), OxbZIP72 and crbzip72 plants were irrigated with 150 mmol/L NaCl solution, a photograph was taken 15 d after the WT displayed extreme chlorosis.B?F, Survival rate (B), relative water content (RWC) (C), total chlorophyll content in the leaf (D), total malondialdehyde (MDA) content (E) and total H2O2 content (F) in WT, OxbZIP72 and crbzip72 plants after salinity stress. Data present Mean ± SE (n = 3). Asterisk denotes significant differences between WT and transgenic lines at P < 0.01 (**) and P < 0.05 (*), respectively as determined by the Student’s t-test.
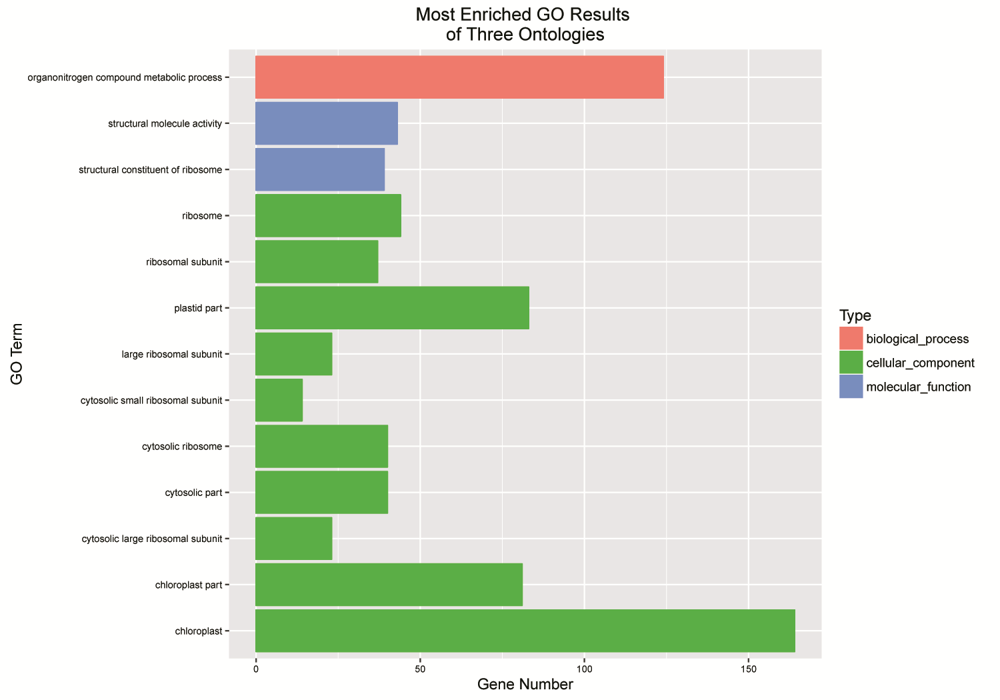
Fig. S2. Gene ontology (GO) investigation of DEGs revealing the barplot of remarkably enriched GO terms. The X-axis denotes genes number belonging to the GO terms on the left. The GO terms are shown on Y-axis.
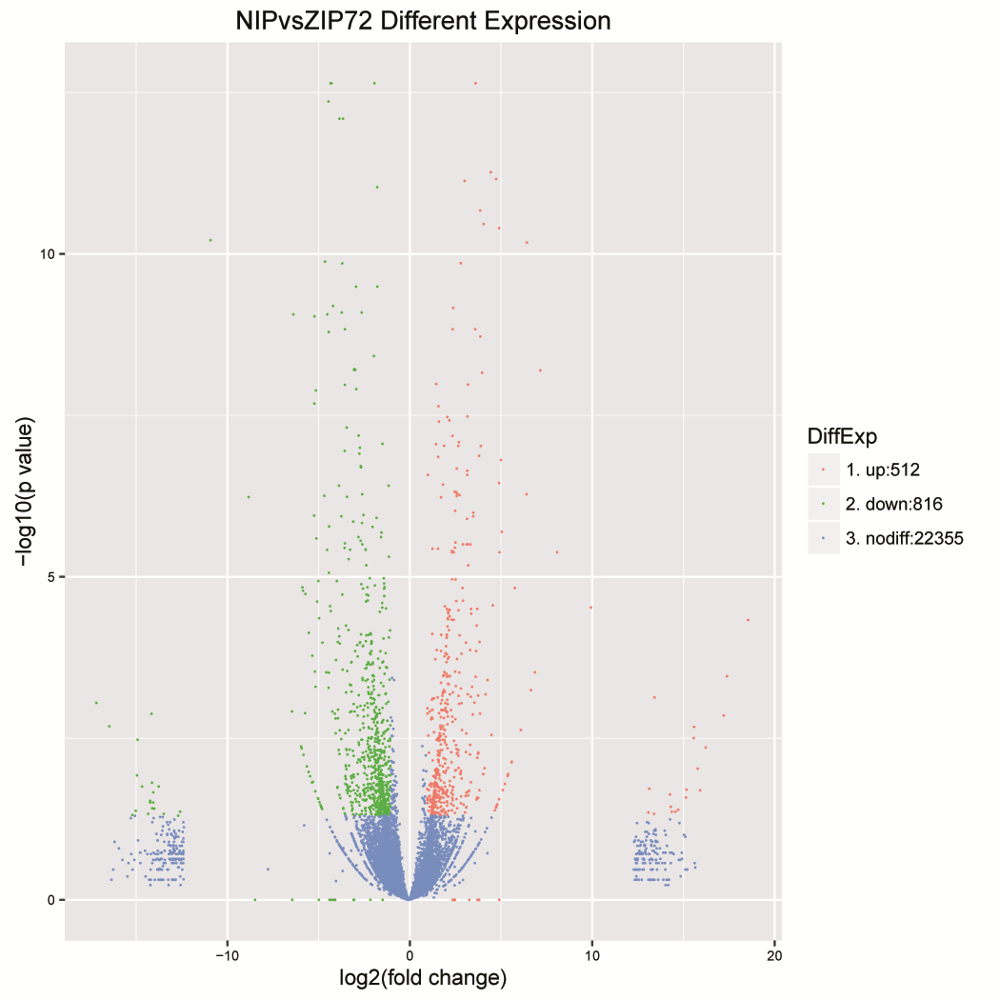
Fig. S3. Volcano plot to visualize the extent of differential expression of genes in this study. The log odds of differential expression were revealed on Y-axis, and x- axis denote the change in log fold.
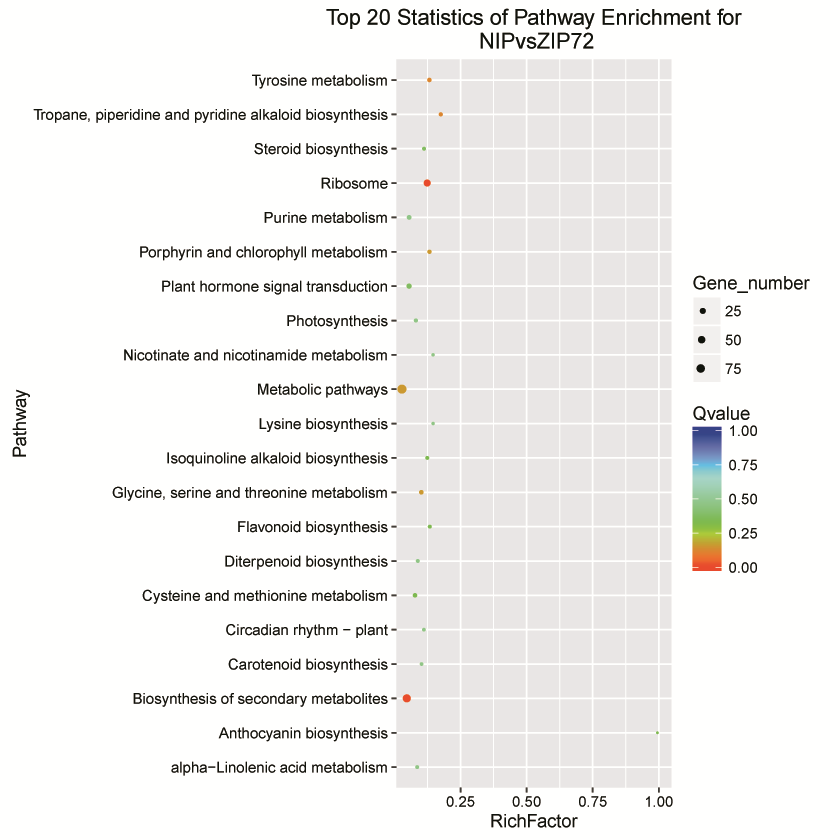
Fig. S4. Molecular interaction network of Genes and Genomes (KEGG pathways). . The ratio of the numbers of enriched DEGs revealed in the KEGG pathway in this study was presented as the rich factor and the values of illustrated underlying genes in this pathway. X-axis revealed the rich factor of each pathway. Y-axis denotes KEGG pathways.
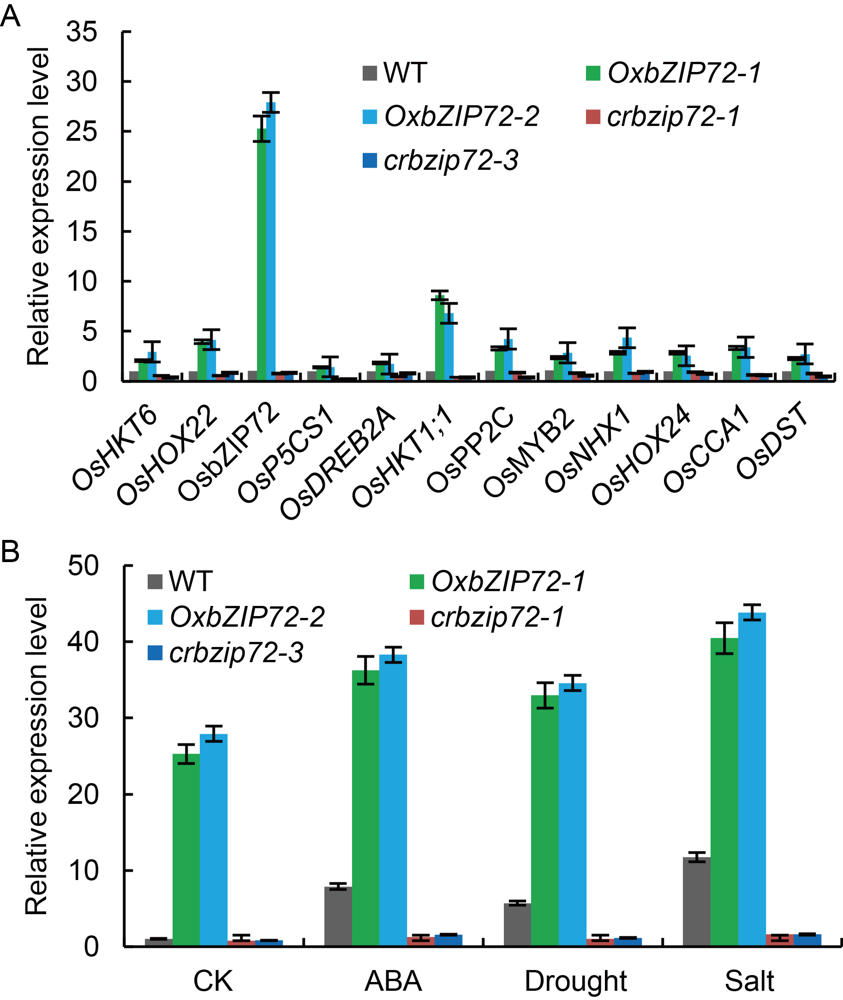
Fig. 4. Transcript of OsbZIP72 regulated genes in OsbZIP72 over- expression (OxbZIP72) and knock-out (crbzip72) transgenic plants by qRT-PCR. A, Differentially expressed gene validation as revealed by RNA-seq exploration and some selected abiotic regulator genes. B, Analysis of transcripts levels of OsbZIP72 in OxbZIP72 and crbzip72 transgenic plants in response to salinity, dehydration and abscisic acid (ABA) by qRT-PCR. Two-week-old seedlings were placed in solution containing 50 µmol/L ABA for 6 h as ABA treatment, 200 mmol/L NaCl for 6 h as salt stress treatment, and for dehydration treatment, seedlings were allowed to air dry in a hood for 5 h at 27 ºC.OsActin1 was used as an internal control. Data are Mean ± SD (n = 3).
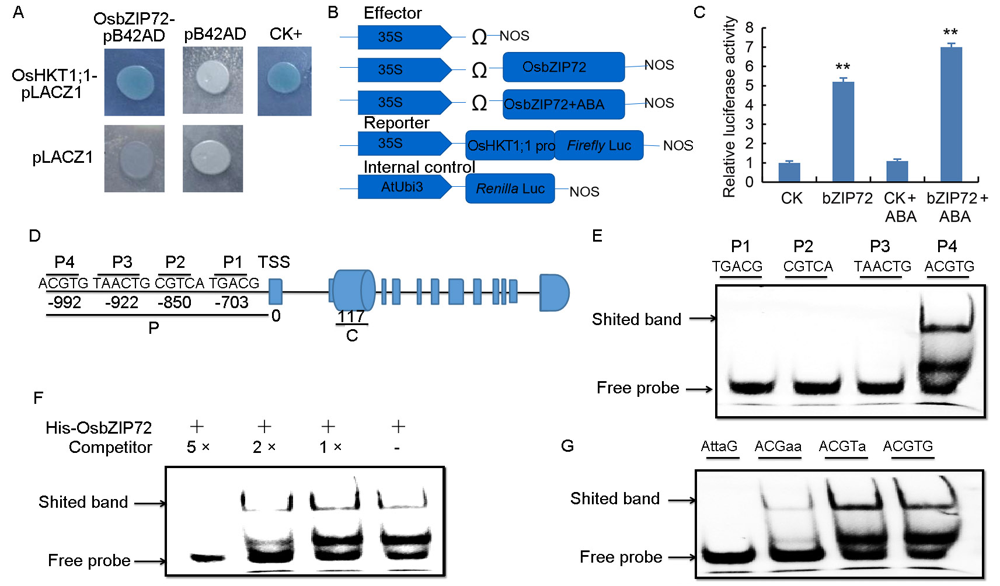
Fig. 5. OsbZIP72 directly binds to abscisic acid responsive element (ABRE) in promoter of OsHKT1;1 and activates its transcription. A, Yeast one-hybrid assay revealing the interaction of OsbZIP72 with the promoter of OsHKT1;1. B and C, Luciferase transcriptional assay using rice protoplasts. AtUbi3:rLUC was used as an internal control. Data are presented as means of three independent replicates and standard errors. ** signifies significant difference at P < 0.01 as determined by the Student’s t-test. D, Position of probes on OsHKT1;1 promoter. Numbers signify the distances (bp) of probes designed to the transcription starting site (TSS), which was set as 0. Black letters indicate the regulatory element sequences in the probe position. E, Electrophoresis mobility shift assay (EMSA) to reveal the binding of OsbZIP72 to P4 in the promoter of OsHKT1;1. F, EMSA showing the binding of OsbZIP72 to ABRE element in P4 in the promoter of OsHKT1;1 in the presence of unlabeled probe as a competitor. G, EMSA revealing the binding of OsbZIP72 to various mutated ABRE elements in P4 in the promoter of OsHKT1;1.
| [1] | Busk P K, Pagès M. 1998. Regulation of abscisic acid-induced transcription. Plant Mol Biol, 37(3): 425‒435. |
| [2] | Chen H, Chen W, Zhou J L, He H, Chen L B, Chen H D, Deng X W. 2012. Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice. Plant Sci, 193: 8‒17. |
| [3] | Deppmann C D, Alvania R S, Taparowsky E J. 2006. Cross- species annotation of basic leucine zipper factor interactions: Insight into the evolution of closed interaction networks. Mol Biol Evol, 23(8): 1480‒1492. |
| [4] | Ellenberger T E, Brandl C J, Struhl K, Harrison S C. 1992. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: Crystal structure of the protein-DNA complex. Cell, 71(7): 1223‒1237. |
| [5] | Foster R, Izawa T, Chua N H. 1994. Plant bZIP proteins gather at ACGT elements. FASEB J, 8(2): 192‒200. |
| [6] | Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez M M, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. 2005. AREB1 is a transcription activator of novel ABRE- dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell, 17(12): 3470‒3488. |
| [7] | Guiltinan M J, Marcotte W R, Quatrano R S. 1990. A plant leucine zipper protein that recognizes an abscisic acid response element. Science, 250: 267‒271. |
| [8] | Guo Z, Ou W, Lu S Y, Zhong Q. 2006. Differential responses of antioxidative system to chilling and drought in four rice cultivars differing in sensitivity. Plant Physiol Biochem, 44(11/12): 828‒836. |
| [9] | Hattori T, Terada T, Hamasuna S. 1995. Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: Analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J, 7(6): 913‒925. |
| [10] | Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J, 6(2): 271‒282. |
| [11] | Hong J Y, Chae M J, Lee I S, Lee Y N, Nam M H, Kim D Y, Byun M O K, Yoon I S. 2011. Phosphorylation-mediated regulation of a rice ABA responsive element binding factor. Phytochemistry, 72(1): 27‒36. |
| [12] | Horie T, Motoda J O, Kubo M, Yang H, Yoda K, Horie R, Chan W Y, Leung H Y, Hattori K, Konomi M. 2005. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J, 44(6): 928‒938. |
| [13] | Horie T, Costa A, Kim T H, Han M J, Horie R, Leung H Y, Miyao A, Hirochika H, An G, Schroeder J I. 2007. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J, 26(12): 3003‒3014. |
| [14] | Hossain M A, Cho J Il, Han M, Ahn C H, Jeon J S, An G, Park P B. 2010a. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol, 167(17): 1512‒1520. |
| [15] | Hossain M A, Lee Y, Cho J Il, Ahn C H, Lee S K, Jeon J S, Kang H, Lee C H, An G, Park P B. 2010b. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol, 72: 557‒566. |
| [16] | Hou Y X, Wang L Y, Wang L, Liu L M, Li L, Sun L, Rao Q, Zhang J, Huang S W. 2015. JMJ704 positively regulates rice defense response against Xanthomonas oryzae pv. oryzae infection via reducing H3K4me2/3 associated with negative disease resistance regulators. BMC Plant Biol, 15(1): 286. |
| [17] | Izawa T, Foster R, Chua N H. 1993. Plant bZIP protein DNA binding specificity. J Mol Biol, 230(4): 1131‒1144. |
| [18] | Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. 2002. bZIP transcription factors in Arabidopsis. Trends Plant Sci, 7(3): 106‒111. |
| [19] | Ji Q, Zhang L S, Wang Y Y, Wang J. 2009. Genome-wide analysis of basic leucine zipper transcription factor families in Arabidopsis thaliana, Oryza sativa and Populus trichocarpa. J Shanghai Univ (Engl Ed), 13(2): 174‒182. |
| [20] | Jin X F, Xiong A S, Peng R H, Liu J G, Gao F, Chen J M, Yao Q H. 2010. OsAREB1, an ABRE-binding protein responding to ABA and glucose, has multiple functions in Arabidopsis. BMB Rep, 43(1): 34‒39. |
| [21] | Kang J Y, Choi H L, Im M Y, Kim S Y. 2002. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell, 14(2): 343‒357. |
| [22] | Landschulz W H, Johnson P F, McKnight S L. 1988. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science, 240: 1759‒1764. |
| [23] | Leng N, Dawson J A, Thomson J A, Ruotti V, Rissman A I, Smits B M G, Haag J D, Gould M N, Stewart R M, Kendziorski C. 2013. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics, 29(8): 1035‒1043. |
| [24] | Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, van de Peer Y, Rouzé P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl Acids Res, 30(1): 325‒327. |
| [25] | Li B, Dewey C N. 2011. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinfo, 12(1): 323. |
| [26] | Liu C T, Wu Y B, Wang X P. 2012. bZIP transcription factor OsbZIP52/RISBZ5: A potential negative regulator of cold and drought stress response in rice. Planta, 235(6): 1157‒1169. |
| [27] | Lopez-Molina L, Mongrand S, Chua N H. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA, 98(8): 4782‒4787. |
| [28] | Lu G J, Gao C X, Zheng X N, Han B. 2009. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta, 229(3): 605‒615. |
| [29] | Ma X L, Zhang Q Y, Zhu Q L, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G. 2015. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant, 8(8): 1274‒1284. |
| [30] | Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn D J, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N. 2002. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett, 531(2): 157‒161. |
| [31] | Møller I S, Gilliham M, Jha D, Mayo G M, Roy S J, Coates J C, Haseloff J, Tester M. 2009. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell, 21(7): 2163‒2178. |
| [32] | Mukherjee K, Choudhury A R, Gupta B, Gupta S, Sengupta D N. 2006. An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of indica rice. BMC Plant Biol, 6(1): 18. |
| [33] | Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annu Rev Plant Biol, 59: 651‒681. |
| [34] | Nakagawa H, Ohmiya K, Hattori T. 1996. A rice bZIP protein, designated OsBZ8, is rapidly induced by abscisic acid. Plant J, 9(2): 217‒227. |
| [35] | Nijhawan A, Jain M, Tyagi A K, Khurana J P. 2008. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol, 146(2): 333‒350. |
| [36] | Oh S J, Song S I k, Kim Y S, Jang H J, Kim S Y, Kim M, Kim Y K, Nahm B H, Kim J K. 2005. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol, 138(1): 341‒351. |
| [37] | Ren Z H, Gao J P, Li L G, Cai X L, Huang W, Chao D Y, Zhu M Z, Wang Z Y, Luan S, Lin H X. 2005. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet, 37(10): 1141‒1146. |
| [38] | Rodriguez-Uribe L, O’Connell M A. 2006. A root-specific bZIP transcription factor is responsive to water deficit stress in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris). J Exp Bot, 57(6): 1391‒1398. |
| [39] | Roychoudhury A, Paul S, Basu S. 2013. Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep, 32(7): 985‒1006. |
| [40] | Seki M, Umezawa T, Urano K, Shinozaki K. 2007. Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol, 10(3): 296‒302. |
| [41] | Shkolnik I D, Adler G, Bar Z D. 2013. ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J, 73(6): 993‒1005. |
| [42] | Skriver K, Olsen F L, Rogers J C, Mundy J. 1991. cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA, 88(16): 7266‒7270. |
| [43] | Tang N, Zhang H, Li X H, Xiao J H, Xiong L Z. 2012. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol, 158(4): 1755‒1768. |
| [44] | Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi S K. 2000. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA, 97(21): 11632‒11637. |
| [45] | Wang R, Jing W, Xiao L Y, Jin Y K, Shen L K, Zhang W H. 2015. The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol, 168(3): 1076‒1090. |
| [46] | Wang T B, Gassmann W, Rubio F, Schroeder J I, Glass A D M. 1998. Rapid up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol, 118(2): 651‒659. |
| [47] | Wang W, Vinocur B, Altman A. 2003. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta, 218(1): 1‒14. |
| [48] | Xiang Y, Tang N, Du H, Ye H Y, Xiong L Z. 2008. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol, 148(4): 1938‒1952. |
| [49] | Xie K B, Yang Y N. 2013. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant, 6(6): 1975‒1983. |
| [50] | Xiong L M, Schumaker K S, Zhu J K. 2002. Cell signaling during cold, drought, and salt stress. Plant Cell, 14: S165‒S183. |
| [51] | Yamaguchi-Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol, 57: 781‒803. |
| [52] | Yang D C, Wu L Y, Hwang Y S, Chen L F, Huang N. 2001. Expression of the REB transcriptional activator in rice grains improves the yield of recombinant proteins whose genes are controlled by a Reb-responsive promoter. Proc Natl Acad Sci USA, 98(20): 11438‒11443. |
| [53] | Yang X, Yang Y N, Xue L J, Zou M J, Liu J Y, Chen F, Xue H W. 2011. Rice ABI5-like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol, 156(3): 1397‒1409. |
| [54] | Zhang J, Nallamilli B R, Mujahid H, Peng Z H. 2010. OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J, 64(4): 604‒617. |
| [55] | Zhu J K. 2002. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol, 53(1): 247‒273. |
| [56] | Zou M J, Guan Y C, Ren H B, Zhang F, Chen F. 2007. Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem Biophys Res Commun, 360(2): 307‒313. |
| [57] | Zou M J, Guan Y C, Ren H B, Zhang F, Chen F. 2008. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol, 66(6): 675‒683. |
| [1] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [2] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [3] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [4] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [5] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [6] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [7] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [8] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [9] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [10] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [11] | Jiang Changjie, Liang Zhengwei, Xie Xianzhi. Priming for Saline-Alkaline Tolerance in Rice: Current Knowledge and Future Challenges [J]. Rice Science, 2023, 30(5): 417-425. |
| [12] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [13] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [14] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| [15] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||