
Rice Science ›› 2024, Vol. 31 ›› Issue (5): 556-571.DOI: 10.1016/j.rsci.2024.06.001
• Research Papers • Previous Articles Next Articles
Kanokwan Kaewmungkun1,2, Keasinee Tongmark2, Sriprapai Chakhonkaen2, Numphet Sangarwut2, Theerachai Thanananta1, Amorntip Muangprom2( )
)
Received:2024-02-16
Accepted:2024-04-18
Online:2024-09-28
Published:2024-10-11
Contact:
Amorntip Muangprom (amorntip.mua@biotec.or.th; amuangprom@gmail.com)
Kanokwan Kaewmungkun, Keasinee Tongmark, Sriprapai Chakhonkaen, Numphet Sangarwut, Theerachai Thanananta, Amorntip Muangprom. Bulked Segregant RNA-Seq Analysis of Pollinated Pistils Reveals Genes Influencing Spikelet Fertility in Rice[J]. Rice Science, 2024, 31(5): 556-571.
Add to citation manager EndNote|Ris|BibTeX
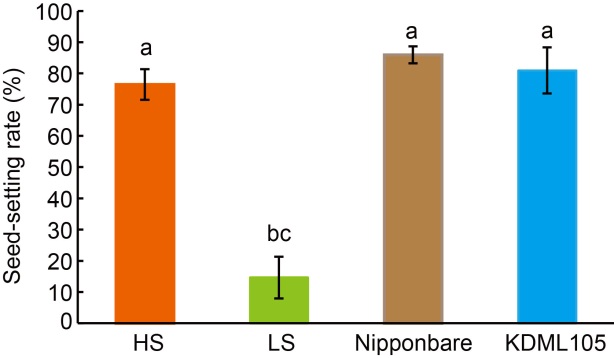
Fig. 1. Rice plants with high (HS) and low seed-setting rates (LS) in F5 population, as well as their parents Nipponbare and KDML105. Data are Mean ± SD (n = 48), and different lowercase letters above the bars show significant differences at P < 0.05.
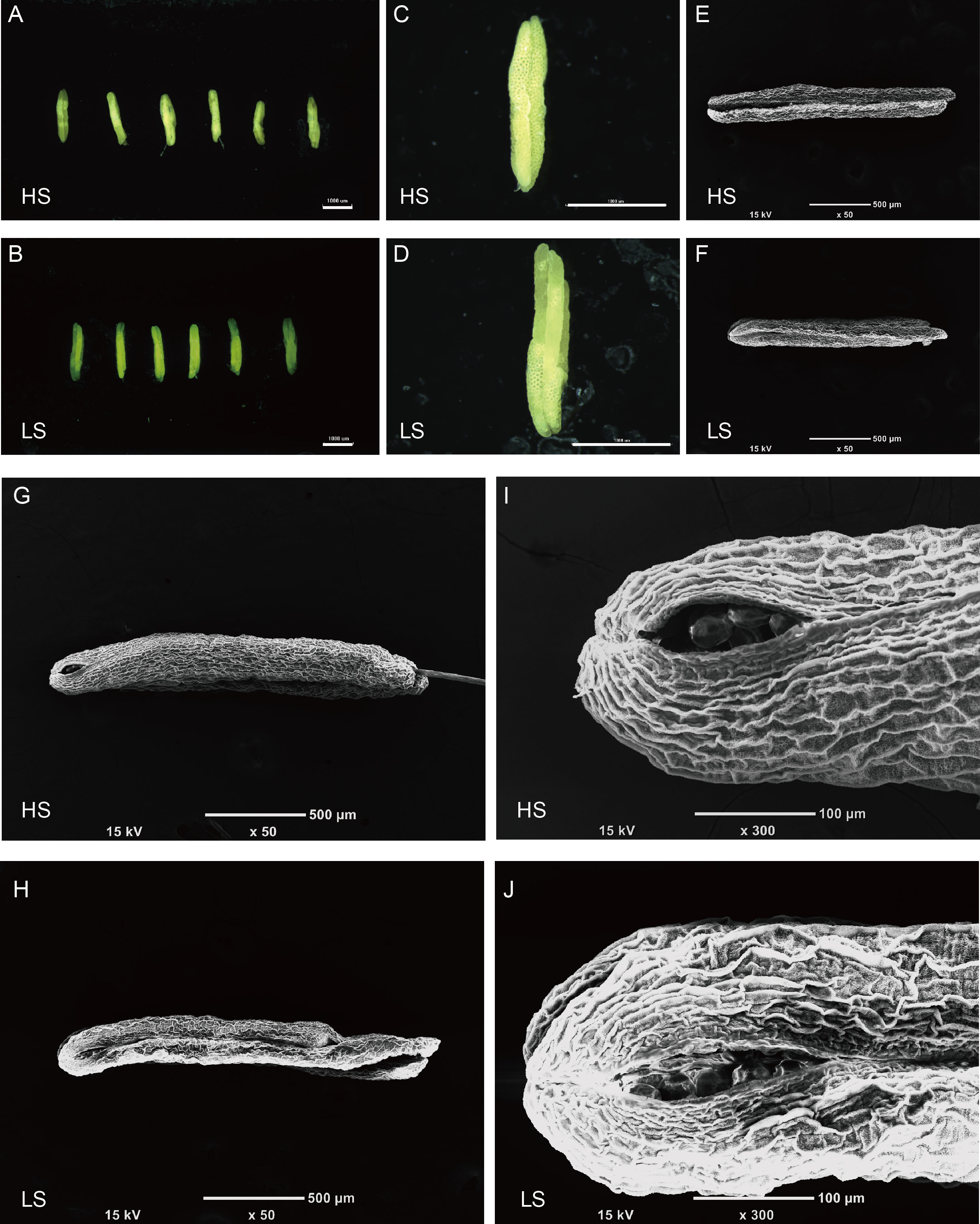
Fig. 2. Microscopic analysis of anther morphology in high (HS) and low seed- setting rate (LS) lines at anthesis in F5 generation. A and B, Morphology of six mature anthers from HS (A) and LS (B) lines. Scale bars are 1 mm, and the field diameters are 6.7×. C and D, Light microscope images of an anther from HS (C) and LS (D) lines. Scale bars are 1 mm, and the field diameters are 30×. E and F, Scanning electron microscope images of an anther from HS (E) and LS (F) lines. Scale bar are 500 µm, and the field diameters are 50×. G and H, Morphology of dehisced anthers from HS (G) and LS (H) lines. Scale bars are 500 µm, and the field diameters are 50×. I and J, Dehiscence for pollen dispersal at the apical of anthers from HS (I) and LS (J) lines. Scale bars are 100 µm, and the field diameters are 300×.

Fig. 3. Filled grains and pollen fertility in high (HS) and low seed-setting rate (LS) rice lines. A and B, Filled grains in a plant (A) and a panicle (B) in HS and LS rice lines. Scale bars are 1 cm. The red arrows represent a zoom from the red square. C, Pollen stained with I2-KI solution in HS and LS rice lines. Scale bars are 200 μm. D, Pollen fertility rate of HS and LS rice lines.
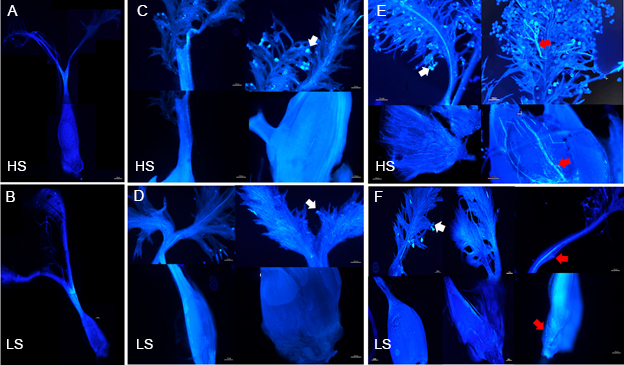
Fig. 4. Pollen-stigma adhesion, pollen grain germination, and pollen tube growth of high (HS) and low seed- setting rate (LS) rice lines in vivo. A and B, Aniline blue staining of HS (A) and LS (B) pistils at 1‒2 h before flowering (BF). C and D, Aniline blue staining of HS (C) and LS (D) pistils at 1‒2 h after flowering with the pollinated state. E and F, Aniline blue staining of HS (E) and LS (F) pistils at 4‒5 h after pollination. Scale bars are 100 μm. The white arrows indicate pollen-stigma adhesion, and the red arrows indicate pollen tubes.
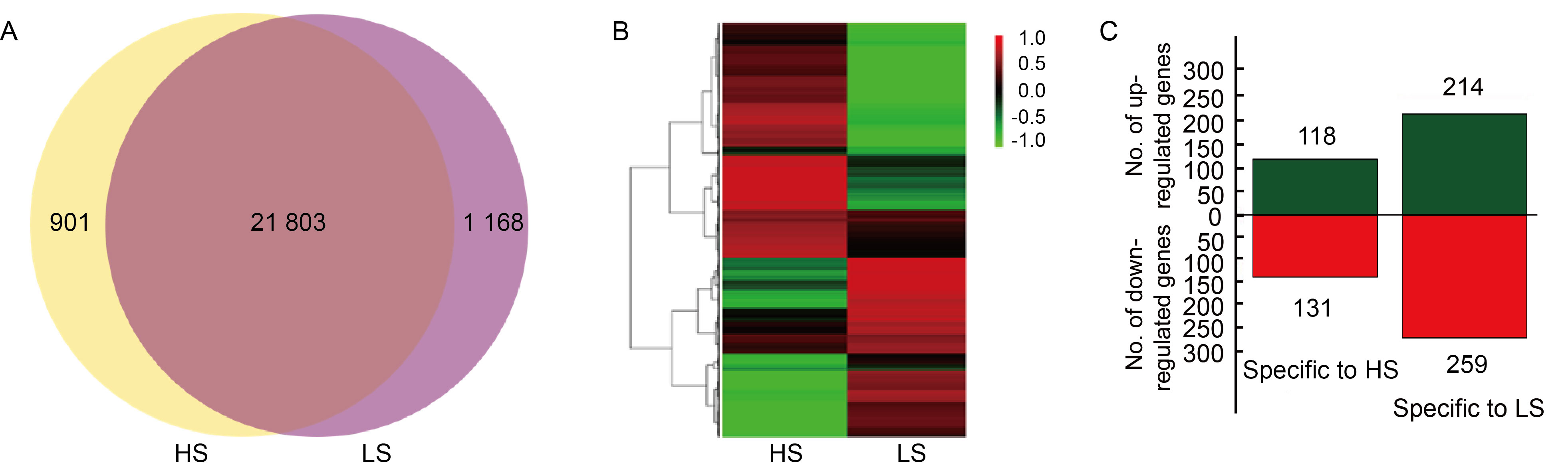
Fig. 5. Co-expression, hierarchical cluster analysis, and significant differentially expressed genes (DEGs) specific to high (HS) and low seed-setting rate (LS) rice lines. A, Venn diagram presenting the number of genes uniquely expressed in each sample, with overlapping regions indicating the number of genes expressed in both samples, based on a 0.5 FPKM (fragments per kilobase of transcript per million reads) threshold. B, Hierarchical clustering of genes specifically expressed in HS and LS samples based on FPKM values. Green indicates lower expression, while red indicates higher expression. Columns represent individual experiments, and rows represent genes. C, The number of up-regulated and down-regulated genes specific to HS and LS rice lines. Green and red represent up-regulated and down-regulated DEGs, respectively, and the numbers on the columns indicate the count of DEGs.
| Gene ID | Gene | Gene annotation | KEGG orthology accession | Log2(Fold change) |
|---|---|---|---|---|
| Os06g0133000 | Wx | Granule-bound starch synthase, synthesis of amylose in endosperm | K13679 | 2.32 |
| Os09g0457400 | Amy3A | Alpha-amylase isozyme 3A precursor (1,4-alpha-d-glucan glucanohydrolase) | K01176 | 2.07 |
| Os06g0185100 | Similar to estradiol 17-beta dehydrogenase 8 | K00059 | 1.94 | |
| Os11g0701100 | Similar to class III chitinase homologue | K01183 | 1.83 | |
| Os02g0467600 | Similar to cinnamate 4-hydroxylase CYP73 | K00487 | 1.82 | |
| Os03g0382100 | Thiolase-like, subgroup domain containing protein | K15397 | 1.45 | |
| Os02g0134400 | Fumarate reductase/succinate dehydrogenase flavoprotein, C-terminal domain containing protein | K00278 | -1.07 | |
| Os03g0670200 | Similar to YJR013Wp (Fragment) | K05284 | -1.20 | |
| Os03g0368900 | Haem peroxidase family protein | K00430 | -1.20 | |
| Os04g0573100 | HTH1 | Putative glucose-methanol-choline oxidoreductase, cutin biosynthesis, anther development, and pollen fertility | K07729 | -1.25 |
| Os03g0321800 | Similar to transcription factor WRKY55 | K23735 | -1.46 |
Table 1. Eleven high seed-setting rate (HS)-specific differentially expressed genes assigned to metabolic pathways using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database.
| Gene ID | Gene | Gene annotation | KEGG orthology accession | Log2(Fold change) |
|---|---|---|---|---|
| Os06g0133000 | Wx | Granule-bound starch synthase, synthesis of amylose in endosperm | K13679 | 2.32 |
| Os09g0457400 | Amy3A | Alpha-amylase isozyme 3A precursor (1,4-alpha-d-glucan glucanohydrolase) | K01176 | 2.07 |
| Os06g0185100 | Similar to estradiol 17-beta dehydrogenase 8 | K00059 | 1.94 | |
| Os11g0701100 | Similar to class III chitinase homologue | K01183 | 1.83 | |
| Os02g0467600 | Similar to cinnamate 4-hydroxylase CYP73 | K00487 | 1.82 | |
| Os03g0382100 | Thiolase-like, subgroup domain containing protein | K15397 | 1.45 | |
| Os02g0134400 | Fumarate reductase/succinate dehydrogenase flavoprotein, C-terminal domain containing protein | K00278 | -1.07 | |
| Os03g0670200 | Similar to YJR013Wp (Fragment) | K05284 | -1.20 | |
| Os03g0368900 | Haem peroxidase family protein | K00430 | -1.20 | |
| Os04g0573100 | HTH1 | Putative glucose-methanol-choline oxidoreductase, cutin biosynthesis, anther development, and pollen fertility | K07729 | -1.25 |
| Os03g0321800 | Similar to transcription factor WRKY55 | K23735 | -1.46 |
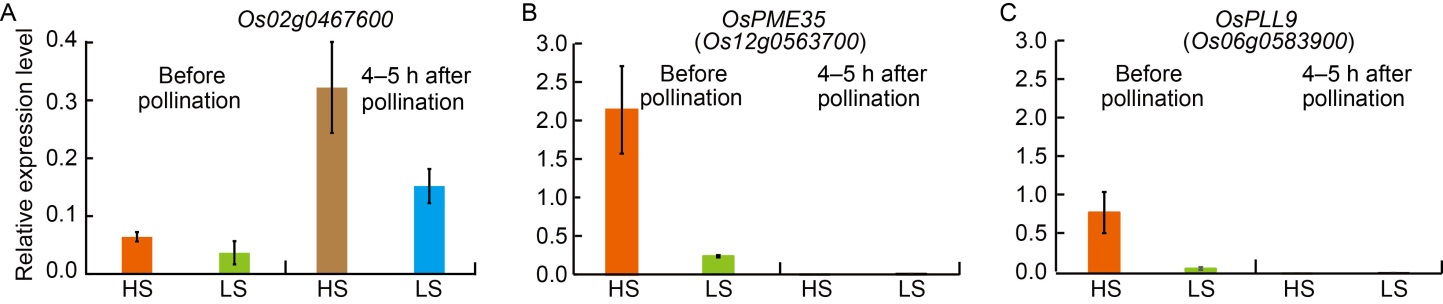
Fig. 6. Expression analysis of high (HS)- and low seed-setting rate (LS)-specific differentially expressed genes (DEGs) before and after pollination by qRT-PCR. A‒C, Relative expression levels of Os02g0467600 (A), Os12g0563700 (B), and Os06g0583900 (C). The relative expression values were normalized to the rice UBQ5 gene (OsUBQ5). The results were obtained from two biological replicates. Error bars indicate standard deviation.
| Gene ID | Gene | Gene annotation | KEGG orthology accession | Log2(Fold change) |
|---|---|---|---|---|
| Pentose and glucuronate interconversion pathway | ||||
| Os03g0300500 | OsPME10 | Pectin methylesterase 10 | K01051 | 1.43 |
| Os12g0563700 | OsPME35 | Pectin lyase fold domain-containing protein | K01051 | 1.13 |
| Os02g0214400 | OsPLL3 | Pectate lyase-like protein | K01728 | 1.21 |
| Os06g0583900 | OsPLL9 | Pectate lyase | K01728 | 1.30 |
| Flavonoid biosynthesis pathway | ||||
| Os02g0767300 | Flavonol synthase/flavanone 3-hydroxylase | K05278 | -1.34 | |
| Os04g0101400 | Flavone synthase II, biosynthesis of tricin O-linked conjugate | K23180 | -1.73 | |
Table 2. Highest fold enrichment of low seed-setting rate (LS)-specific differentially expressed genes (DEGs) in pentose and glucuronate interconversion and flavonoid biosynthesis pathways.
| Gene ID | Gene | Gene annotation | KEGG orthology accession | Log2(Fold change) |
|---|---|---|---|---|
| Pentose and glucuronate interconversion pathway | ||||
| Os03g0300500 | OsPME10 | Pectin methylesterase 10 | K01051 | 1.43 |
| Os12g0563700 | OsPME35 | Pectin lyase fold domain-containing protein | K01051 | 1.13 |
| Os02g0214400 | OsPLL3 | Pectate lyase-like protein | K01728 | 1.21 |
| Os06g0583900 | OsPLL9 | Pectate lyase | K01728 | 1.30 |
| Flavonoid biosynthesis pathway | ||||
| Os02g0767300 | Flavonol synthase/flavanone 3-hydroxylase | K05278 | -1.34 | |
| Os04g0101400 | Flavone synthase II, biosynthesis of tricin O-linked conjugate | K23180 | -1.73 | |
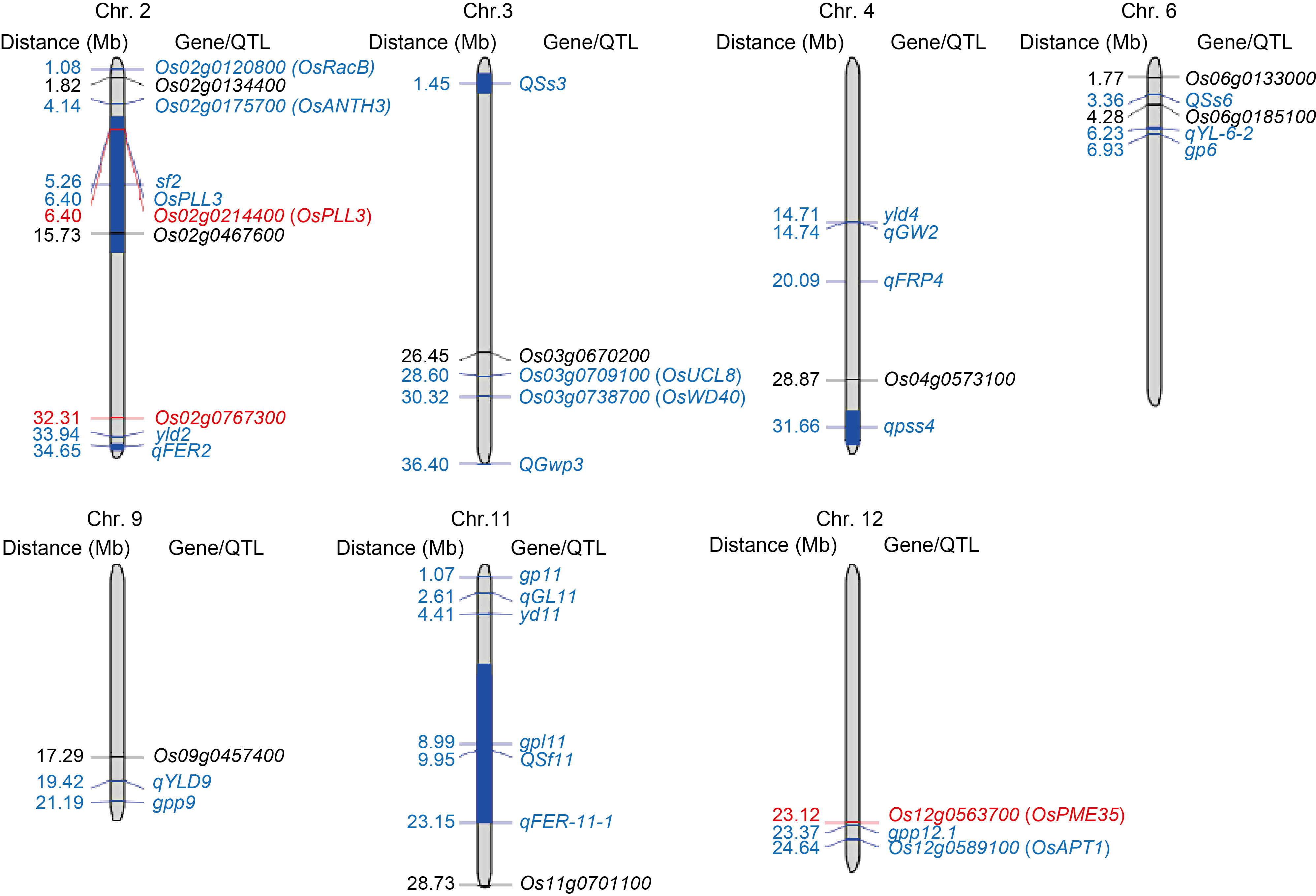
Fig. 7. Physical maps showing co-localization of dominant enrichment genes specific to high (HS) and low seed-setting rates (LS) with previously identified QTLs/genes involved in spikelet fertility. The previously identified QTLs and functional genes involved in spikelet fertility, pollen growth, and yield components are labeled in blue, and the dominant enrichment differentially expressed genes specific to HS and LS are labeled in black and red, respectively.
| [1] |
Alqudah A M, Sharma R, Börner A. 2021. Insight into the genetic contribution of maximum yield potential, spikelet development and abortion in barley. Plants People Planet, 3(6): 721-736.
DOI |
| [2] |
Becker J D, Takeda S, Borges F, Dolan L, Feijó J A. 2014. Transcriptional profiling of Arabidopsis root hairs and pollen defines an apical cell growth signature. BMC Plant Biol, 14: 197.
DOI PMID |
| [3] | Benitez L C, da Maia L C, Ribeiro M V, Pegoraro C, Peters J A, de Oliveira A C, de M J Ariano M, Braga E J B. 2013. Salt induced change of gene expression in salt sensitive and tolerant rice species. J Agric Sci, 5(10): 251-260. |
| [4] |
Birchler J A. 2015. Heterosis: The genetic basis of hybrid vigour. Nat Plants, 1: 15020.
DOI PMID |
| [5] |
Bosch M, Hepler P K. 2005. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell, 17(12): 3219-3226.
DOI PMID |
| [6] |
Chao J T, Li Z Y, Sun Y H, Aluko O O, Wu X R, Wang Q, Liu G S. 2021. MG2C: A user-friendly online tool for drawing genetic maps. Mol Hortic, 1(1): 16.
DOI PMID |
| [7] | Chari J, Wilson P. 2001. Factors limiting hybridization between Penstemon spectabilis and Penstemon centranthifolius. Can J Bot, 79(12): 1439-1448. |
| [8] |
Chen H Q, Zhang Z G, Ni E D, Lin J W, Peng G Q, Huang J L, Zhu L Y, Deng L, Yang F F, Luo Q, Sun W, Liu Z L, Zhuang C X, Liu Y G, Zhou H. 2020. HMS1 interacts with HMS1I to regulate very-long-chain fatty acid biosynthesis and the humidity-sensitive genic male sterility in rice (Oryza sativa). New Phytol, 225(5): 2077-2093.
DOI PMID |
| [9] |
Chen H Y, Zhao Z G, Liu L L, Kong W Y, Lin Y, You S M, Bai W T, Xiao Y J, Zheng H, Jiang L, Li J, Zhou J W, Tao D Y, Wan J M. 2017. Genetic analysis of a hybrid sterility gene that causes both pollen and embryo sac sterility in hybrids between Oryza sativa L. and Oryza longistaminata. Heredity, 119(3): 166-173.
DOI PMID |
| [10] | Chen L, Bian J M, Shi S L, Yu J F, Khanzada H, Wassan G M, Zhu C L, Luo X, Tong S, Yang X R, Peng X S, Yong S, Yu Q Y, He X P, Fu J R, Chen X R, Hu L F, Ouyang L J, He H H. 2018. Genetic analysis for the grain number heterosis of a super-hybrid rice WFYT025 combination using RNA-Seq. Rice, 11(1): 37. |
| [11] | Chen L, Yuan Y, Wu J W, Chen Z X, Wang L, Shahid M Q, Liu X D. 2019. Carbohydrate metabolism and fertility related genes high expression levels promote heterosis in autotetraploid rice harboring double neutral genes. Rice, 12(1): 34. |
| [12] | Chen T T, Ma J Y, Xu C M, Jiang N, Li G Y, Fu W M, Feng B H, Wang D Y, Wu Z H, Tao L X, Fu G F. 2022. Increased ATPase activity promotes heat-resistance, high-yield, and high-quality traits in rice by improving energy status. Front Plant Sci, 13: 1035027. |
| [13] | Conze L L, Berlin S, Le Bail A, Kost B. 2017. Transcriptome profiling of tobacco (Nicotiana tabacum) pollen and pollen tubes. BMC Genomics, 18(1): 581. |
| [14] | Distefano G, Gentile A, Herrero M. 2011. Pollen-pistil interactions and early fruiting in parthenocarpic citrus. Ann Bot, 108(3): 499-509. |
| [15] | El-Maarouf-Bouteau H. 2022. The seed and the metabolism regulation. Biology, 11(2): 168. |
| [16] |
Fernández-González M, González-Fernández E, Fernández-González D, Rodríguez-Rajo F J. 2020. Secondary outcomes of the Ole e 1 proteins involved in pollen tube development: Impact on allergies. Front Plant Sci, 11: 974.
DOI PMID |
| [17] | Foreman J, Demidchik V, Bothwell J H F, Mylona P, Miedema H, Torres M A, Linstead P, Costa S, Brownlee C, Jones J D G, Davies J M, Dolan L. 2003. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature, 422: 442-446. |
| [18] | Fu D H, Xiao M L, Hayward A, Fu Y, Liu G, Jiang G J, Zhang H H. 2014. Utilization of crop heterosis: A review. Euphytica, 197(2): 161-173. |
| [19] |
Fujii S, Tsuchimatsu T, Kimura Y, Ishida S, Tangpranomkorn S, Shimosato-Asano H, Iwano M, Furukawa S, Itoyama W, Wada Y, Shimizu K K, Takayama S. 2019. A stigmatic gene confers interspecies incompatibility in the Brassicaceae. Nat Plants, 5(7): 731-741.
DOI PMID |
| [20] | Gao Y B, Du L H, Ma Q, Yuan Y H, Liu J R, Song H, Feng B L. 2022. Conjunctive analyses of bulk segregant analysis sequencing and bulk segregant RNA sequencing to identify candidate genes controlling spikelet sterility of foxtail millet. Front Plant Sci, 13: 842336. |
| [21] |
Ge S X, Jung D, Yao R N. 2020. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics, 36(8): 2628-2629.
DOI PMID |
| [22] | Gournas C, Papageorgiou I, Diallinas G. 2008. The nucleobase- ascorbate transporter (NAT) family: Genomics, evolution, structure- function relationships and physiological role. Mol Biosyst, 4(5): 404-416. |
| [23] | Guo J, Xu X M, Li W T, Zhu W Y, Zhu H T, Liu Z Q, Luan X, Dai Z J, Liu G F, Zhang Z M, Zeng R Z, Tang G, Fu X L, Wang S K, Zhang G Q. 2016. Overcoming inter-subspecific hybrid sterility in rice by developing indica-compatible japonica lines. Sci Rep, 6: 26878. |
| [24] | Guo J X, Liu Y G. 2012. Molecular control of male reproductive development and pollen fertility in rice. J Integr Plant Biol, 54(12): 967-978. |
| [25] |
Hasegawa K, Kamada S, Takehara S, Takeuchi H, Nakamura A, Satoh S, Iwai H. 2020. Rice putative methyltransferase gene OsPMT16 is required for pistil development involving pectin modification. Front Plant Sci, 11: 475.
DOI PMID |
| [26] |
Hennig L, Gruissem W, Grossniklaus U, Köhler C. 2004. Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol, 135(3): 1765-1775.
DOI PMID |
| [27] | Hu L F, Liang W Q, Yin C S, Cui X, Zong J, Wang X, Hu J P, Zhang D B. 2011. Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell, 23(2): 515-533. |
| [28] |
Huang J Y, Zhao X B, Cheng K, Jiang Y H, Ouyang Y D, Xu C G, Li X H, Xiao J H, Zhang Q F. 2013. OsAP65, a rice aspartic protease, is essential for male fertility and plays a role in pollen germination and pollen tube growth. J Exp Bot, 64(11): 3351-3360.
DOI PMID |
| [29] | Huang W J, Liu H K, McCormick S, Tang W H. 2014. Tomato pistil factor STIG1 promotes in vivo pollen tube growth by binding to phosphatidylinositol 3-phosphate and the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell, 26( 6): 2505-2523. |
| [30] | Huang X R, Peng X B, Sun M X. 2017. OsGCD1 is essential for rice fertility and required for embryo dorsal-ventral pattern formation and endosperm development. New Phytol, 215(3): 1039-1058. |
| [31] | Initiative I B. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature, 463: 763-768. |
| [32] | International Rice Genome Sequencing Project. 2005. The map-based sequence of the rice genome. Nature, 436: 793-800. |
| [33] |
Jiang L X, Yang S L, Xie L F, Puah C S, Zhang X Q, Yang W C, Sundaresan V, Ye D. 2005. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell, 17(2): 584-596.
DOI PMID |
| [34] | Kamara N, Jiao Y M, Lu Z J, Aloryi K D, Wu J W, Liu X D, Shahid M Q. 2021. Cytological observations and bulked-segregant analysis coupled global genome sequencing reveal two genes associated with pollen fertility in tetraploid rice. Int J Mol Sci, 22(2): 841. |
| [35] | Khlaimongkhon S, Chakhonkaen S, Tongmark K, Sangarwut N, Panyawut N, Wasinanon T, Sikaewtung K, Wanchana S, Mongkolsiriwatana C, Chunwonges J, Muangprom A. 2021. RNA sequencing reveals rice genes involved in male reproductive development under temperature alteration. Plants, 10(4): 663. |
| [36] |
Khush G S. 2005. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol Biol, 59(1): 1-6.
DOI PMID |
| [37] | Kim E J, Park S W, Hong W J, Silva J, Liang W Q, Zhang D B, Jung K H, Kim Y J. 2020. Genome-wide analysis of RopGEF gene family to identify genes contributing to pollen tube growth in rice (Oryza sativa). BMC Plant Biol, 20(1): 95. |
| [38] | Kim E J, Kim J H, Hong W J, Kim E Y, Kim M H, Lee S K, Min C W, Kim S T, Park S K, Jung K H, Kim Y J. 2023. Rice pollen- specific OsRALF17 and OsRALF19 are essential for pollen tube growth. J Integr Plant Biol, 65(9): 2218-2236. |
| [39] | Kim M J, Jeon B W, Oh E, Seo P J, Kim J. 2021. Peptide signaling during plant reproduction. Trends Plant Sci, 26(8): 822-835. |
| [40] | Kim Y J, Jeong H Y, Kang S Y, Silva J, Kim E J, Park S K, Jung K H, Lee C H. 2020. Physiological importance of pectin modifying genes during rice pollen development. Int J Mol Sci, 21(14): 4840. |
| [41] | Kovaleva L V, Zakharova E V. 2004. Gametophyte-sporophyte interactions in the pollen-pistil system: 4. The hormonal status and the mechanism of self-incompatibility. Russ J Plant Physiol, 51(3): 402-406. |
| [42] | Kwon C T, Kim S H, Kim D, Paek N C. 2015. The rice floral repressor Early flowering1affects spikelet fertility by modulating gibberellin signaling. Rice, 8(1): 58. |
| [43] | Lee S K, Hong W J, Silva J, Kim E J, Park S K, Jung K H, Kim Y J. 2021. Global identification of ANTH genes involved in rice pollen germination and functional characterization of a key member, OsANTH3. Front Plant Sci, 12: 609473. |
| [44] |
Leroux C, Bouton S, Kiefer-Meyer M C, Fabrice T N, Mareck A, Guénin S, Fournet F, Ringli C, Pelloux J, Driouich A, Lerouge P, Lehner A, Mollet J C. 2015. PECTIN METHYLESTERASE48 is involved in Arabidopsis pollen grain germination. Plant Physiol, 167(2): 367-380.
DOI PMID |
| [45] | Li S C, Li W B, Huang B, Cao X M, Zhou X Y, Ye S M, Li C B, Gao F Y, Zou T, Xie K L, Ren Y, Ai P, Tang Y F, Li X M, Deng Q M, Wang S Q, Zheng A P, Zhu J, Liu H N, Wang L X, Li P. 2013. Natural variation in PTB1 regulates rice seed setting rate by controlling pollen tube growth. Nat Commun, 4: 2793. |
| [46] | Liu K, Chen Y, Huang J, Qiu Y Y, Li S Y, Zhuo X X, Yu F, Gao J, Li G M, Zhang W Y, Zhang H, Gu J F, Liu L J, Yang J C. 2022. Spikelet differentiation and degeneration in rice varieties with different panicle sizes. Food Energy Secur, 11(1): e320. |
| [47] | Liu S S, Zhong J, Ling S, Liu Y Q, Xu Y, Yao J L. 2021. OsAPT1 a pollen preferentially expressed gene is essential for pollen tube germination and elongation in rice. Plant Mol Biol Rep, 39(1): 87-97. |
| [48] | Liu Z H, Li S, Li W, Liu Q, Zhang L L, Song X Y. 2020. Comparative transcriptome analysis indicates that a core transcriptional network mediates isonuclear alloplasmic male sterility in wheat (Triticum aestivum L.). BMC Plant Biol, 20(1): 10. |
| [49] | Majeed A, Johar P, Raina A, Salgotra R K, Feng X Z, Bhat J A. 2022. Harnessing the potential of bulk segregant analysis sequencing and its related approaches in crop breeding. Front Genet, 13: 944501. |
| [50] |
Marioni J C, Mason C E, Mane S M, Stephens M, Gilad Y. 2008. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res, 18(9): 1509-1517.
DOI PMID |
| [51] | Mazuecos-Aguilera I, Suárez-Santiago V N. 2023. Identification of candidate genes involved in the determinism of pollen grain aperture morphology by comparative transcriptome analysis in Papaveraceae. Plants, 12(7): 1570. |
| [52] | Moran Lauter A N, Muszynski M G, Huffman R D, Scott M P. 2017. A pectin methylesterase ZmPme3 is expressed in Gametophyte factor1-s (Ga1-s) silks and maps to that locus in maize (Zea mays L.). Front Plant Sci, 8: 1926. |
| [53] | Nadir S, Khan S, Zhu Q, Henry D, Wei L, Lee D S, Chen L J. 2018. An overview on reproductive isolation in Oryza sativa complex. AoB Plants, 10(6): ply060. |
| [54] |
Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science, 320: 1344-1349.
DOI PMID |
| [55] |
Nasrallah J B. 2023. Stop and go signals at the stigma-pollen interface of the Brassicaceae. Plant Physiol, 193(2): 927-948.
DOI PMID |
| [56] | Negi M S, Devic M, Delseny M, Lakshmikumaran M. 2000. Identification of AFLP fragments linked to seed coat colour in Brassica juncea and conversion to a SCAR marker for rapid selection. Theor Appl Genet, 101(1): 146-152. |
| [57] | Nevhudzholi K M, Gwata E T, McHau G R A. 2020. A genetic study of pod shattering resistance in F2 soybean (Glycine max) germplasm. S Afr J Plant Soil, 37(2): 174-176. |
| [58] | Nguyen H P, Jeong H Y, Kim H, Kim Y C, Lee C H. 2016. Molecular and biochemical characterization of rice pectin methylesterase inhibitors (OsPMEIs). Plant Physiol Biochem, 101: 105-112. |
| [59] | Niu S S, Yu Y M, Xu C G, Li G W, Ouyang Y D. 2014. Prezygotic reproductive isolation and fertility in crosses between indica and japonica subspecies. Sci Sin Vitae, 44(8): 815-821. (in Chinese with English abstract) |
| [60] | Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. 2009. ATTED- II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res, 37: D987-D991. |
| [61] | Oka H I. 2012. Origin of Cultivated Rice. Amsterdam, the Newsland: Elsevier. |
| [62] |
Ouyang Y D, Huang X L, Lu Z H, Yao J L. 2012. Genomic survey, expression profile and co-expression network analysis of OsWD40 family in rice. BMC Genomics, 13: 100.
DOI PMID |
| [63] | Park J R, Kim E G, Jang Y H, Kim K M. 2021. Screening and identification of genes affecting grain quality and spikelet fertility during high-temperature treatment in grain filling stage of rice. BMC Plant Biol, 21(1): 263. |
| [64] | Park M, Lee J H, Han K, Jang S, Han J, Lim J H, Jung J W, Kang B C. 2019. A major QTL and candidate genes for capsaicinoid biosynthesis in the pericarp of Capsicum chinense revealed using QTL-seq and RNA-seq. Theor Appl Genet, 132(2): 515-529. |
| [65] | Parre E, Geitmann A. 2005. Pectin and the role of the physical properties of the cell wall in pollen tube growth of Solanum chacoense. Planta, 220(4): 582-592. |
| [66] |
Paterson A H, Bowers J E, Chapman B A. 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA, 101(26): 9903-9908.
DOI PMID |
| [67] |
Potocký M, Jones M A, Bezvoda R, Smirnoff N, Žárský V. 2007. Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phytol, 174(4): 742-751.
DOI PMID |
| [68] | Ramsey J, Bradshaw Jr H D, Schemske D W. 2003. Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae). Evolution, 57(7): 1520-1534. |
| [69] |
Robinson M D, McCarthy D J, Smyth G K. 2010. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1): 139-140.
DOI PMID |
| [70] | Scholz P, Anstatt J, Krawczyk H E, Ischebeck T. 2020. Signalling pinpointed to the tip: The complex regulatory network that allows pollen tube growth. Plants, 9(9): 1098. |
| [71] | Selinski J, Scheibe R. 2014. Pollen tube growth: Where does the energy come from? Plant Signal Behav, 9(12): e977200. |
| [72] | Sera Y, Hanamata S, Sakamoto S, Ono S, Kaneko K, Mitsui Y, Koyano T, Fujita N, Sasou A, Masumura T, Saji H, Nonomura K I, Mitsuda N, Mitsui T, Kurusu T, Kuchitsu K. 2019. Essential roles of autophagy in metabolic regulation in endosperm development during rice seed maturation. Sci Rep, 9(1): 18544. |
| [73] | Shahid M Q, Sun J F, Wei C M, Zhang P, Liu X D. 2010. Studies on the abnormality of embryo sac and pollen fertility in autotetraploid rice during different growing seasons. Pak J Bot, 42(1): 7-9. |
| [74] |
Shi Z, Ren W, Zhao Y X, Wang X Q, Zhang R Y, Su A G, Wang S, Li C H, Wang J R, Wang S S, Zhang Y X, Ji Y L, Song W, Zhao J R. 2021. Identification of a locus associated with genic male sterility in maize via EMS mutagenesis and bulked-segregant RNA-seq. Crop J, 9(6): 1263-1269.
DOI |
| [75] | Tao D Y, McNally K L, Koide Y, Matsubara K. 2021. Editorial: Reproductive barriers and gene introgression in rice species. Front Plant Sci, 12: 699761. |
| [76] | Tang L, Xu Z J, Chen W F. 2017. Advances and prospects of super rice breeding in China. J Integr Agric, 16(5): 984-991. |
| [77] | Tian G W, Chen M H, Zaltsman A, Citovsky V. 2006. Pollen- specific pectin methylesterase involved in pollen tube growth. Dev Biol, 294(1): 83-91. |
| [78] | Tran L T, Sugimoto K, Kasozi M, Mitalo O W, Ezura H. 2023. Pollination, pollen tube growth, and fertilization independently contribute to fruit set and development in tomato. Front Plant Sci, 14: 1205816. |
| [79] | Wang L D, Filatov D A. 2023. Mechanisms of prezygotic post- pollination reproductive barriers in plants. Front Plant Sci, 14: 1230278. |
| [80] | Wang W X, Li Y Y, Dang P Q, Zhao S J, Lai D W, Zhou L G. 2018. Rice secondary metabolites: Structures, roles, biosynthesis, and metabolic regulation. Molecules, 23(12): 3098. |
| [81] |
Wang Z Y, Zheng F Q, Shen G Z, Gao J P, Snustad D P, Li M G, Zhang J L, Hong M M. 1995. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J, 7(4): 613-622.
DOI PMID |
| [82] | Warman C, Panda K, Vejlupkova Z, Hokin S, Unger-Wallace E, Cole R A, Chettoor A M, Jiang D, Vollbrecht E, Evans M M S, Slotkin R K, Fowler J E. 2020. High expression in maize pollen correlates with genetic contributions to pollen fitness as well as with coordinated transcription from neighboring transposable elements. PLoS Genet, 16(4): e1008462. |
| [83] |
Widmer A, Lexer C, Cozzolino S. 2009. Evolution of reproductive isolation in plants. Heredity, 102(1): 31-38.
DOI PMID |
| [84] |
Wu J W, Shahid M Q, Guo H B, Yin W, Chen Z X, Wang L, Liu X D, Lu Y G. 2014. Comparative cytological and transcriptomic analysis of pollen development in autotetraploid and diploid rice. Plant Reprod, 27(4): 181-196.
DOI PMID |
| [85] | Xiang X J, Zhang P P, Yu P, Zhang Y X, Yang Z F, Sun L P, Wu W X, Khan R M, Abbas A, Cheng S H, Cao L Y. 2019. LSSR1 facilitates seed setting rate by promoting fertilization in rice. Rice, 12(1): 31. |
| [86] | Xu Y, Yang J, Wang Y H, Wang J C, Yu Y, Long Y, Wang Y L, Zhang H, Ren Y L, Chen J, Wang Y, Zhang X, Guo X P, Wu F Q, Zhu S S, Lin Q B, Jiang L, Wu C Y, Wang H Y, Wan J M. 2017a. OsCNGC13 promotes seed-setting rate by facilitating pollen tube growth in stylar tissues. PLoS Genet, 13(7): e1006906. |
| [87] | Xu Y, Liu S S, Liu Y Q, Ling S, Chen C S, Yao J L. 2017b. HOTHEAD-like HTH1 is involved in anther cutin biosynthesis and is required for pollen fertility in rice. Plant Cell Physiol, 58(7): 1238-1248. |
| [88] | Xu Y F, Cai W G, Chen X F, Chen M J, Liang W Q. 2022. A small Rho GTPase OsRacB is required for pollen germination in rice. Dev Growth Differ, 64(2): 88-97. |
| [89] | Xue Z Y, Xu X, Zhou Y, Wang X N, Zhang Y C, Liu D, Zhao B B, Duan L X, Qi X Q. 2018. Deficiency of a triterpene pathway results in humidity-sensitive genic male sterility in rice. Nat Commun, 9(1): 604. |
| [90] | Yang J C, Zhou Y J, Jiang Y. 2022. Amino acids in rice grains and their regulation by polyamines and phytohormones. Plants, 11(12): 1581. |
| [91] | Yu B, Liu L T, Wang T. 2019. Deficiency of very long chain alkanes biosynthesis causes humidity-sensitive male sterility via affecting pollen adhesion and hydration in rice. Plant Cell Environ, 42(12): 3340-3354. |
| [92] | Zhang C X, Li G Y, Chen T T, Feng B H, Fu W M, Yan J X, Islam M R, Jin Q Y, Tao L X, Fu G F. 2018. Heat stress induces spikelet sterility in rice at anthesis through inhibition of pollen tube elongation interfering with auxin homeostasis in pollinated pistils. Rice, 11(1): 14. |
| [93] | Zhang F, Zhang Y C, Zhang J P, Yu Y, Zhou Y F, Feng Y Z, Yang Y W, Lei M Q, He H, Lian J P, Chen Y Q. 2018. Rice UCL8, a plantacyanin gene targeted by miR408, regulates fertility by controlling pollen tube germination and growth. Rice, 11(1): 60. |
| [94] | Zhang H Y, Kjemtrup-Lovelace S, Li C B, Luo Y, Chen L P, Song B H. 2017. Comparative RNA-seq analysis uncovers a complex regulatory network for soybean cyst nematode resistance in wild soybean (Glycine soja). Sci Rep, 7(1): 9699. |
| [95] |
Zhang K, Song Q, Wei Q, Wang C C, Zhang L W, Xu W Y, Su Z. 2016. Down-regulation of OsSPX1 caused semi-male sterility, resulting in reduction of grain yield in rice. Plant Biotechnol J, 14(8): 1661-1672.
DOI PMID |
| [96] | Zhang Z, Sun W, Wen L Y, Liu Y Q, Guo X L, Liu Y, Yao C S, Xue Q W, Sun Z C, Wang Z M, Zhang Y H. 2023. Dynamic gene regulatory networks improving spike fertility through regulation of floret primordia fate in wheat. Plant Cell Environ, 46(11): 3628-3643. |
| [97] | Zhang Z G, Zhang B C, Chen Z B, Zhang D M, Zhang H R, Wang H, Zhang Y E, Cai D R, Liu J, Xiao S L, Huo Y Q, Liu J, Zhang L J, Wang M M, Liu X, Xue Y B, Zhao L, Zhou Y H, Chen H B. 2018. A PECTIN METHYLESTERASE gene at the maize Ga1 locus confers male function in unilateral cross-incompatibility. Nat Commun, 9(1): 3678. |
| [98] | Zheng Y Z, Yan J J, Wang S Z, Xu M L, Huang K K, Chen G L, Ding Y. 2018. Genome-wide identification of the pectate lyase-like (PLL) gene family and functional analysis of two PLL genes in rice. Mol Genet Genomics, 293(6): 1317-1331. |
| [99] | Zhou L, Wang Y Z, Xu X B, Yan D, Yu W J, Miao Y F, Xu B. 2022. Conjunctive analyses of BSA-seq and BSR-seq unveil the Msβ-GAL and MsJMT as key candidate genes for cytoplasmic male sterility in alfalfa (Medicago sativa L.). Int J Mol Sci, 23(13): 7172. |
| [1] | Wu Qi, Tao Ye, Zhang Xiaolong, Dong Xiaoying, Xia Jixing, Shen Renfang, Zhu Xiaofang. Pectin Methylesterases Enhance Root Cell Wall Phosphorus Remobilization in Rice [J]. Rice Science, 2022, 29(2): 179-188. |
| [2] | Cheabu Sulaiman, Panichawong Nat, Rattanametta Prisana, Wasuri Boonthong, Kasemsap Poonpipope, Arikit Siwaret, Vanavichit Apichart, Malumpong Chanate. Screening for Spikelet Fertility and Validation of Heat Tolerance in a Large Rice Mutant Population [J]. Rice Science, 2019, 26(4): 229-238. |
| [3] | Yaliang Wang, Lei Wang, Jianxia Zhou, Shengbo Hu, Huizhe Chen, Jing Xiang, Yikai Zhang, Yongjun Zeng, Qinghua Shi, Defeng Zhu, Yuping Zhang. Research Progress on Heat Stress of Rice at Flowering Stage [J]. Rice Science, 2019, 26(1): 1-10. |
| [4] | Cheabu Sulaiman, Moung-ngam Peerapon, Arikit Siwaret, Vanavichit Apichart, Malumpong Chanate. Effects of Heat Stress at Vegetative and Reproductive Stages on Spikelet Fertility [J]. Rice Science, 2018, 25(4): 218-226. |
| [5] | Vivitha P., Raveendran M., Vijayalakshmi D.. Introgression of QTLs Controlling Spikelet Fertility Maintains Membrane Integrity and Grain Yield in Improved White Ponni Derived Progenies Exposed to Heat Stress [J]. Rice Science, 2017, 24(1): 32-40. |
| [6] | Xia Xu, Xiao-bo Zhang, Yong-feng Shi, Hui-mei Wang, Bao-hua Feng, Xiao-hong Li, Qi-na Huang, Li-xin Song, Dan Guo, Yan He, Jian-li Wu. A Point Mutation in an F-Box Domain-Containing Protein Is Responsible for Brown Hull Phenotype in Rice [J]. Rice Science, 2016, 23(1): 1-8. |
| [7] | Liu Wen-qiang, Fan Ye-yang, Chen Jie, Shi Yong-feng, Wu Jian-li. Avoidance of Linkage Drag Between Blast Resistance Gene and the QTL Conditioning Spikelet Fertility Based on Genotype Selection Against Heading Date in Rice [J]. RICE SCIENCE, 2009, 16(1): 21-26 . |
| [8] | FU Guan-fu, TAO Long-xing, SONG Jian, WANG Xi, CAO Li-yong, CHENG Shi-hua. Responses of Yield Characteristics to High Temperature During Flowering Stage in Hybrid Rice Guodao 6 [J]. RICE SCIENCE, 2008, 15(3): 215-222 . |
| [9] | XU Fu-rong, YU Teng-qiong, YAN Hong-mei, TANG Cui-feng, A Xin-xiang, DAI Lu-yuan. Specific Spikelet Fertility as an Indicator of Cold Tolerance Identification at Booting Stage in Rice [J]. RICE SCIENCE, 2006, 13(3): 211-217 . |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||