
Rice Science ›› 2023, Vol. 30 ›› Issue (6): 613-631.DOI: 10.1016/j.rsci.2023.04.007
• Research Papers • Previous Articles Next Articles
Prathap V1, Suresh Kumar1( ), Nand Lal Meena1,2, Chirag Maheshwari1, Monika Dalal3, Aruna Tyagi1(
), Nand Lal Meena1,2, Chirag Maheshwari1, Monika Dalal3, Aruna Tyagi1( )
)
Received:2023-02-10
Accepted:2023-04-27
Online:2023-11-28
Published:2023-08-10
Contact:
Aruna TYAGI (arunatyagi19@yahoo.com);
Suresh KUMAR (sureshkumar@iari.res.in)
Prathap V, Suresh Kumar, Nand Lal Meena, Chirag Maheshwari, Monika Dalal, Aruna Tyagi. Phosphorus Starvation Tolerance in Rice Through Combined Physiological, Biochemical, and Proteome Analyses[J]. Rice Science, 2023, 30(6): 613-631.
Add to citation manager EndNote|Ris|BibTeX
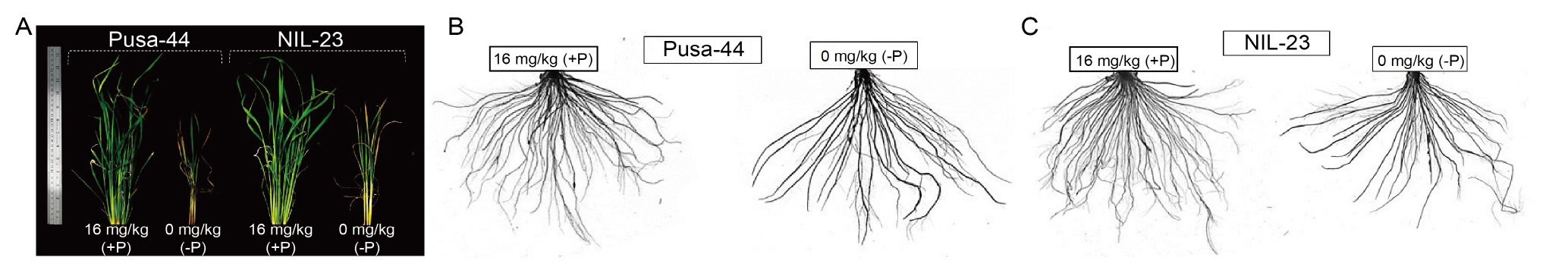
Fig. 1. Shoot and root morphologies of rice genotypes grown hydroponically under P-starvation stress for 30 d. A, Shoot morphology of Pusa-44 (P-sensitive genotype) and NIL-23 (P-tolerant genotype) grown under contrasting P conditions [16 mg/kg (+P) and 0 mg/kg (-P)]. B and C, Root morphology of Pusa-44 (B) and NIL-23 (C) grown under contrasting P conditions [16 mg/kg (+P) and 0 mg/kg (-P)].
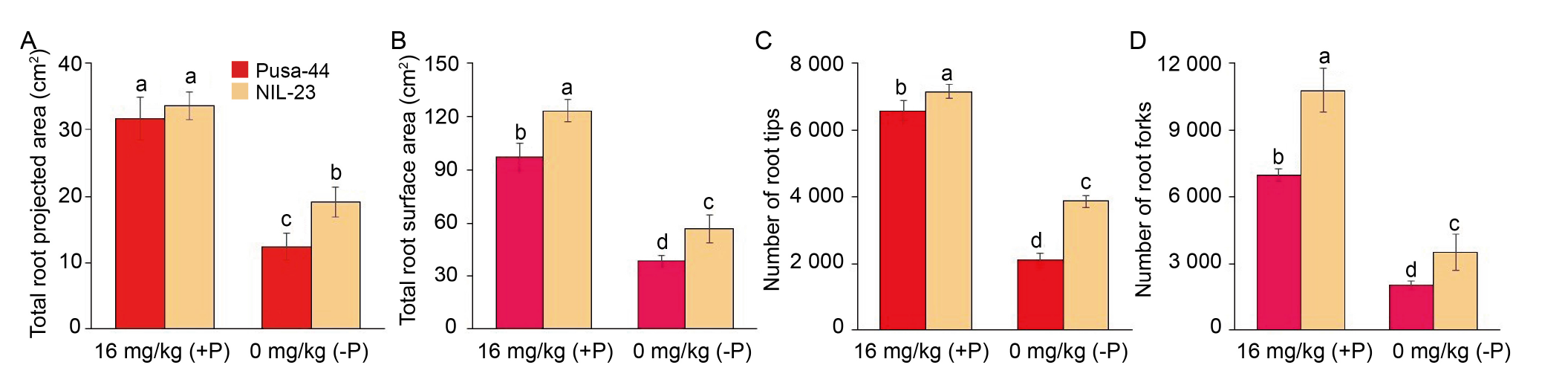
Fig. 2. Modulation in root system architecture of rice genotypes grown hydroponically under P-starvation stress for 30 d. The variations of total root projected area (A), total root surface area (B), the number of root tips (C), and the number of root forks (D) observed in Pusa-44 (P-sensitive genotype) and NIL-23 (P-tolerant genotype) treated with different P concentrations. Data are Mean ± SD (n = 3), and different lowercase letters above the bars show significant differences at P < 0.05.
| Genotype | Growth condition | Total leaf area (cm2) | Total chlorophyll content (mg/g) | Shoot biomass (g) | Root biomass (g) | Root-to-shoot biomass ratio | Shoot length (cm) | Total root length (cm) |
|---|---|---|---|---|---|---|---|---|
| Pusa-44 | -P (0 mg/kg) | 44.96 ± 3.35 c | 12 ± 1.25 c | 0.45 ± 0.10 d | 0.20 ± 0.02 d | 0.45 ± 0.02 b | 22.1 ± 0.9 b | 320.0 ± 14.6 d |
| +P (16 mg/kg) | 182.43 ± 7.30 a | 31 ± 2.65 a | 1.49 ± 0.35 b | 0.53 ± 0.03 b | 0.36 ± 0.03 d | 32.9 ± 2.9 a | 824.7 ± 36.1 b | |
| NIL-23 | -P (0 mg/kg) | 45.71 ± 11.40 c | 17 ± 3.01 b | 0.54 ± 0.20 c | 0.43 ± 0.03 c | 0.80 ± 0.01 a | 24.7 ± 2.3 b | 502.0 ± 88.9 c |
| +P (16 mg/kg) | 168.29 ± 5.40 b | 34 ± 1.98 a | 1.57 ± 0.19 a | 0.63 ± 0.02 a | 0.40 ± 0.02 c | 31.6 ± 1.1 a | 1 103.0 ± 12.5 a |
Table 1. Morpho-physiological variations in rice genotypes [Pusa-44 (P-sensitive genotype) and NIL-23 (P-tolerant genotype)] grown hydroponically under P-starvation stress for 30 d.
| Genotype | Growth condition | Total leaf area (cm2) | Total chlorophyll content (mg/g) | Shoot biomass (g) | Root biomass (g) | Root-to-shoot biomass ratio | Shoot length (cm) | Total root length (cm) |
|---|---|---|---|---|---|---|---|---|
| Pusa-44 | -P (0 mg/kg) | 44.96 ± 3.35 c | 12 ± 1.25 c | 0.45 ± 0.10 d | 0.20 ± 0.02 d | 0.45 ± 0.02 b | 22.1 ± 0.9 b | 320.0 ± 14.6 d |
| +P (16 mg/kg) | 182.43 ± 7.30 a | 31 ± 2.65 a | 1.49 ± 0.35 b | 0.53 ± 0.03 b | 0.36 ± 0.03 d | 32.9 ± 2.9 a | 824.7 ± 36.1 b | |
| NIL-23 | -P (0 mg/kg) | 45.71 ± 11.40 c | 17 ± 3.01 b | 0.54 ± 0.20 c | 0.43 ± 0.03 c | 0.80 ± 0.01 a | 24.7 ± 2.3 b | 502.0 ± 88.9 c |
| +P (16 mg/kg) | 168.29 ± 5.40 b | 34 ± 1.98 a | 1.57 ± 0.19 a | 0.63 ± 0.02 a | 0.40 ± 0.02 c | 31.6 ± 1.1 a | 1 103.0 ± 12.5 a |
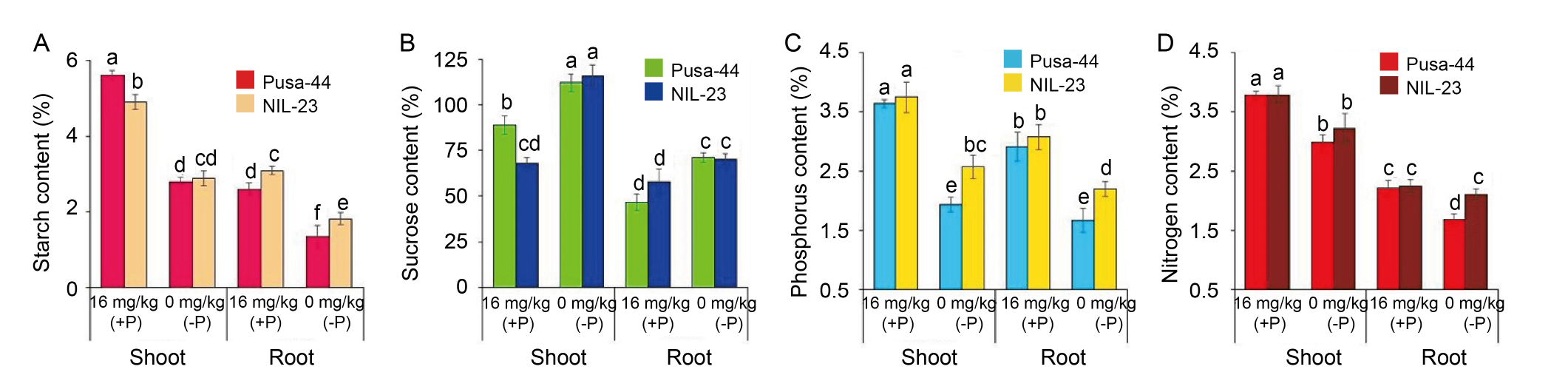
Fig. 3. Effects of P-starvation stress on biochemical parameters in rice genotypes grown hydroponically under P-starvation stress. Effect of the stress on starch content (A), sucrose content (B), phosphorus content (C), and nitrogen content (D) observed in Pusa-44 (the P-sensitive genotype) and NIL-23 (the P-tolerant genotype) grown under 16 mg/kg (+P) and 0 mg/kg (-P) conditions. Data are Mean ± SD (n = 3), and different lowercase letters above the bars show significant differences at P < 0.05.
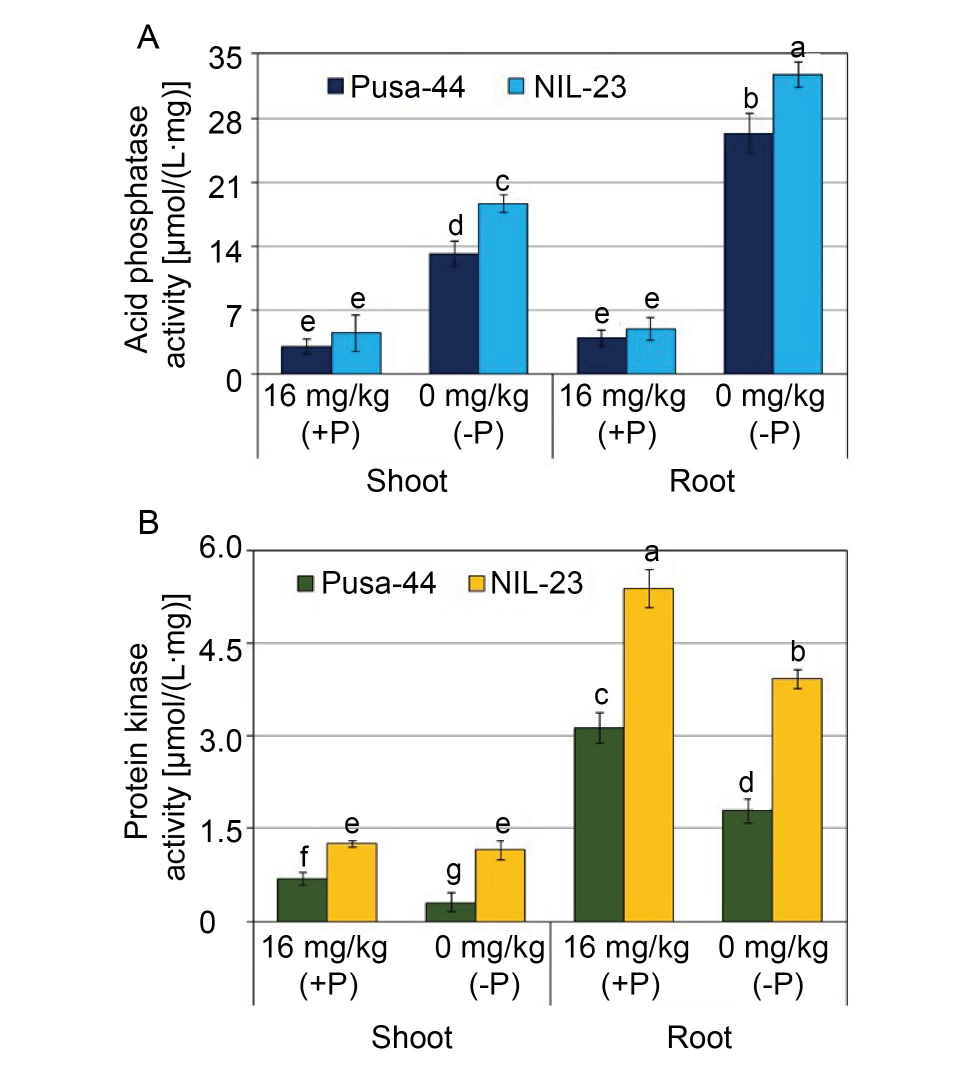
Fig. 4. Effects of P-starvation stress on acid phosphatase (A) and total protein kinase activities (B) in rice genotypes grown hydroponically under P-starvation stress for 30 d. Data are Mean ± SD (n = 3), and different lowercase letters show significant differences at P < 0.05.
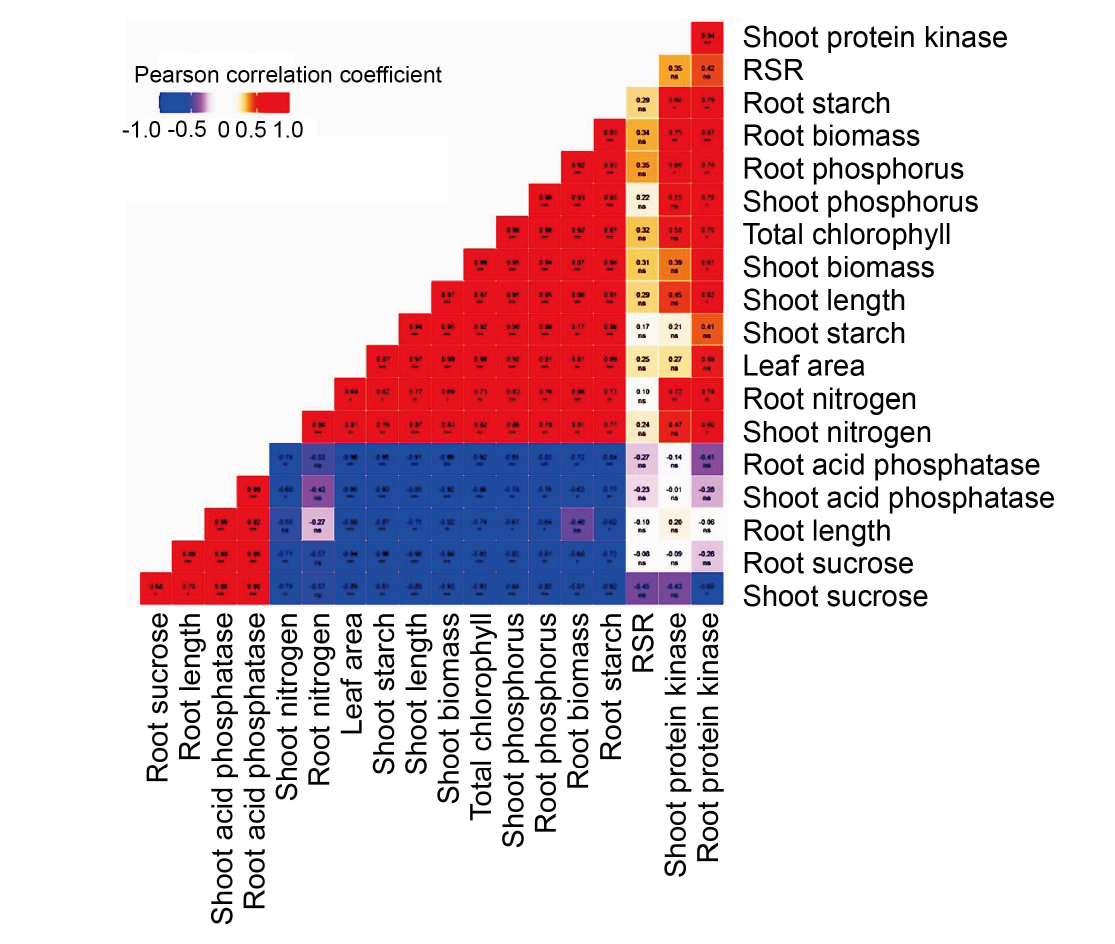
Fig. 5. Pearson correlation (PC) analysis depicting correlation coefficient and an association between physiological and biochemical characteristics. PC coefficient varies between -1 to +1, and a positive linear relationship between variables is highlighted in red and a negative correlation is highlighted in blue. RSR, Root to shoot biomass ratio. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns indicates not significant.
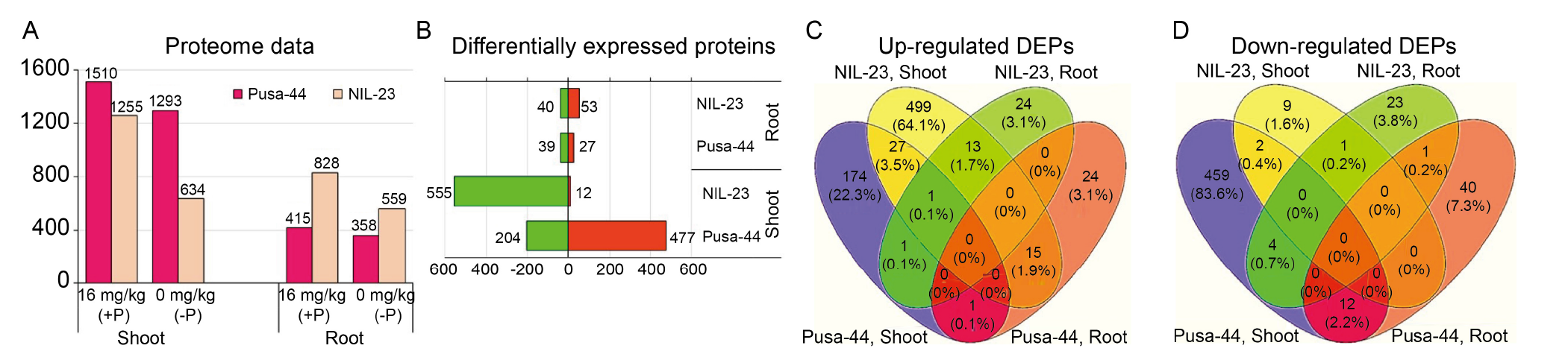
Fig. 6. Comparative analysis of differentially expressed proteins (DEPs) in shoot and root of in rice genotypes [ Pusa-44 (P-sensitive genotype) and NIL-23 (P-tolerant genotype)] under a most contrasting condition [16 mg/kg (+P) and 0 mg/kg (-P)]. A, Number of proteins identified in shoot and root. B, Statistically significant total-, up- (in green) and down-regulated (in red) DEPs. C and D, Four-way Venn diagram showing the significantly up-regulated (C) and down-regulated DEPs (D) in shoot and root.
| [1] | Ågren G I, Wetterstedt J Å M, Billberger M F K. 2012. Nutrient limitation on terrestrial plant growth: Modeling the interaction between nitrogen and phosphorus. New Phytol, 194(4): 953-960. |
| [2] | Alloush G A. 2003. Responses of hydroponically-grown chickpea to low phosphorus: pH changes, nutrient uptake rates, and root morphological changes. Agronomie, 23(2): 123-133. |
| [3] | Assuero S G, Mollier A, Pellerin S. 2004. The decrease in growth of phosphorus-deficient maize leaves is related to a lower cell production. Plant Cell Environ, 27(7): 887-895. |
| [4] | Aziz T, Sabir M, Farooq M, Maqsood M A, Ahmad H R, Warraich, E A. 2014. Phosphorus deficiency in plants: Responses, adaptive mechanisms, and signaling. In: Hakeem K, Rehman R, Tahir I. Plant Signaling: Understanding the Molecular Crosstalk. New Delhi, India: Springer: 133-148. |
| [5] | Bai J H, Xie Y M, Shi M H, Yao S F, Lu W J, Xiao K. 2022. TaMPK2B, a member of the MAPK family in T. aestivum, enhances plant low-Pi stress tolerance through modulating physiological processes associated with phosphorus starvation defensiveness. Plant Sci, 323: 111375. |
| [6] | Bozzo G G, Plaxton W C. 2008. The role of intracellular and secreted purple acid phosphatases in tomato phosphate nutrition. In: Preedy V R, Watson R R. Tomatoes and Tomato Products: Nutritional, Medicinal and Therapeutic Properties. Enfield, USA: Science Publishers, Inc.: 215-234. |
| [7] | Bradford J M. 1976. Partial revision of the Acartia subgenus acartiura (Copepoda: Calanoida: Acartiidae). N Z J Mar Freshw Res, 10(1): 159-202. |
| [8] | Bustos R, Castrillo G, Linhares F, Puga M I, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. 2010. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet, 6(9): e1001102. |
| [9] | Cai H M, Xie W B, Lian X M. 2013. Comparative analysis of differentially expressed genes in rice under nitrogen and phosphorus starvation stress conditions. Plant Mol Biol Rep, 31(1): 160-173. |
| [10] | Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L. 2008. Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot, 59(9): 2479-2497. |
| [11] | Cerana M, Bonza M C, Harris R, Sanders D, Michelis M I. 2006. Abscisic acid stimulates the expression of two isoforms of plasma membrane Ca2+-ATPase in Arabidopsis thaliana seedlings. Plant Biol, 8(5): 572-578. |
| [12] | Cerutti T, Delatorre C A. 2013. Nitrogen and phosphorus interaction and cytokinin: Responses of the primary root of Arabidopsis thaliana and the pdr1 mutant. Plant Sci, 198: 91-97. |
| [13] | Chen J Y, Wang Y F, Wang F, Yang J, Gao M X, Li C Y, Liu Y Y, Liu Y, Yamaji N, Ma J F, Paz-Ares J, Nussaume L, Zhang S Q, Yi K K, Wu Z C, Wu P. 2015. The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell, 27(3): 711-723. |
| [14] | Cheng L, Min W L, Li M, Zhou L L, Hsu C C, Yang X L, Jiang X, Ruan Z J, Zhong Y J, Wang Z Y, Wang W F. 2021. Quantitative proteomics reveals that GmENO2 proteins are involved in response to phosphate starvation in the leaves of Glycine max L. Int J Mol Sci, 22(2): 920. |
| [15] | Chin J H, Lu X C, Haefele S M, Gamuyao R, Ismail A, Wissuwa M, Heuer S. 2010. Development and application of gene-based markers for the major rice QTL Phosphorus uptake 1. Theor Appl Genet, 120(6): 1073-1086. |
| [16] | Chiou T J, Lin S I. 2011. Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol, 62: 185-206. |
| [17] | Clegg K M. 1956. The application of the anthrone reagent to the estimation of starch in cereals. J Sci Food Agric, 7(1): 40-44. |
| [18] | Cook F R, Fahy B, Trafford K. 2012. A rice mutant lacking a large subunit of ADP-glucose pyrophosphorylase has drastically reduced starch content in the culm but normal plant morphology and yield. Funct Plant Biol, 39(12): 1068-1078. |
| [19] | Das S, Tyagi W, Rai M, Yumnam J S. 2017. Understanding Fe2+ toxicity and P deficiency tolerance in rice for enhancing productivity under acidic soils. Biotechnol Genet Eng Rev, 33(1): 97-117. |
| [20] | Deng Q W, Dai L F, Chen Y L, Wu D C, Shen Y, Xie J K, Luo X D. 2022. Identification of phosphorus stress related proteins in the seedlings of Dongxiang wild rice (Oryza rufipogon griff.) using label-free quantitative proteomic analysis. Genes, 13(1): 108. |
| [21] | Ding W L, Cong W F, Lambers H. 2021. Plant phosphorus- acquisition and -use strategies affect soil carbon cycling. Trends Ecol Evol, 36(10): 899-906. |
| [22] | Dissanayaka D M S B, Maruyama H, Nishida S, Tawaraya K, Wasaki J. 2017. Landrace of japonica rice, Akamai exhibits enhanced root growth and efficient leaf phosphorus remobilization in response to limited phosphorus availability. Plant Soil, 414(1): 327-338. |
| [23] | Dissanayaka D M S B, Plaxton W C, Lambers H, Siebers M, Marambe B, Wasaki J. 2018. Molecular mechanisms underpinning phosphorus-use efficiency in rice. Plant Cell Environ, 41(7): 1483-1496. |
| [24] | Finley J, Fellers D A. 1973. Sucrose determination by a modified anthrone method: Application with sweetened wheat-soy blend and corn-soy-milk. Cereal Chem, 50: 210-214. |
| [25] | Föhse D, Claassen N, Jungk A. 1991. Phosphorus efficiency of plants: II. Significance of root radius, root hairs and cation-anion balance for phosphorus influx in seven plant species. Plant Soil, 132(2): 261-272. |
| [26] | Fragoso S, Espíndola L, Páez-Valencia J, Gamboa A, Camacho Y, Martínez-Barajas E, Coello P. 2009. SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol, 149(4): 1906-1916. |
| [27] | Fredeen A L, Raab T K, Rao I M, Terry N. 1990. Effects of phosphorus nutrition on photosynthesis in Glycine max (L.) Merr. Planta, 181(3): 399-405. |
| [28] | Gamuyao R, Chin J H, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza E M, Wissuwa M, Heuer S. 2012. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature, 488: 535-539. |
| [29] | Gu M, Chen A Q, Sun S B, Xu G H. 2016. Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: What is missing? Mol Plant, 9(3): 396-416. |
| [30] | Hallama M, Pekrun C, Lambers H, Kandeler E. 2019. Hidden miners: The roles of cover crops and soil microorganisms in phosphorus cycling through agroecosystems. Plant Soil, 434: 7-45. |
| [31] | Hammond J P, Broadley M R, White P J. 2004. Genetic responses to phosphorus deficiency. Ann Bot, 94(3): 323-332. |
| [32] | Hanson W C. 1950. The photometric determination of phosphorus in fertilizers using the phosphovanado-molybdate complex. J Sci Food Agric, 1(6): 172-173. |
| [33] | Harrison J A, Bouwman A F, Mayorga E, Seitzinger S. 2010. Magnitudes and sources of dissolved inorganic phosphorus inputs to surface fresh waters and the coastal zone: A new global model. Global Biogeochem Cycles, 24(1): GB1003. |
| [34] | He C J, Morgan P W, Drew M C. 1992. Enhanced sensitivity to ethylene in nitrogen- or phosphate-starved roots of Zea mays L. during aerenchyma formation. Plant Physiol, 98(1): 137-142. |
| [35] | Hermans C, Hammond J P, White P J, Verbruggen N. 2006. How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci, 11(12): 610-617. |
| [36] | Hinsinger P, Betencourt E, Bernard L, Brauman A, Plassard C, Shen J B, Tang X Y, Zhang F S. 2011. P for two, sharing a scarce resource: Soil phosphorus acquisition in the rhizosphere of intercropped species. Plant Physiol, 156(3): 1078-1086. |
| [37] | Hodge A, Berta G, Doussan C, Merchan F, Crespi M. 2009. Plant root growth, architecture and function. Plant Soil, 321(1): 153-187. |
| [38] | Holford I C R. 1997. Soil phosphorus: Its measurement, and its uptake by plants. Soil Res, 35(2): 227-240. |
| [39] | Hu B, Chu C C. 2020. Nitrogen-phosphorus interplay: Old story with molecular tale. New Phytol, 225(4): 1455-1460. |
| [40] | Hu B, Jiang Z M, Wang W, Qiu Y H, Zhang Z H, Liu Y Q, Li A F, Gao X K, Liu L C, Qian Y W, Huang X H, Yu F F, Kang S, Wang Y Q, Xie J P, Cao S Y, Zhang L H, Wang Y C, Xie Q, Kopriva S, Chu C C. 2019. Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat Plants, 5(4): 401-413. |
| [41] | Irfan M, Aziz T, Maqsood M A, Bilal H M, Siddique K H M, Xu M G. 2020. Phosphorus (P) use efficiency in rice is linked to tissue-specific biomass and P allocation patterns. Sci Rep, 10: 4278. |
| [42] | Jacob J, Lawlor D W. 1993. Extreme phosphate deficiency decreases the in vivo CO2/O2 specificity factor of ribulose 1, 5-bisphosphate carboxylase-oxygenase in intact leaves of sunflower. J Exp Bot, 44(11): 1635-1641. |
| [43] | Jin J, Tang C X, Armstrong R, Sale P. 2012. Phosphorus supply enhances the response of legumes to elevated CO2 (FACE) in a phosphorus-deficient vertisol. Plant Soil, 358(1): 91-104. |
| [44] | Johnson C B, Holloway B R, Smith H, Grierson D. 1973. Isoenzymes of acid phosphatase in germinating peas. Planta, 115(1): 1-10. |
| [45] | Kafle A, Cope K, Raths R, Krishna Yakha J, Subramanian S, Bücking H, Garcia K. 2019. Harnessing soil microbes to improve plant phosphate efficiency in cropping systems. Agronomy, 9(3): 127. |
| [46] | Kant S, Peng M S, Rothstein S J. 2011. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet, 7(3): e1002021. |
| [47] | Karthikeyan A S, Varadarajan D K, Jain A, Held M A, Carpita N C, Raghothama K G. 2007. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta, 225(4): 907-918. |
| [48] | Kaur S, Seem K, Duhan N, Kumar S, Kaundal R, Mohapatra T. 2023. Transcriptome and physio-biochemical profiling reveals differential responses of rice cultivars at reproductive-stage drought stress. Int J Mol Sci, 24(2): 1002. |
| [49] | Kavanová M, Alfredo Lattanzi F, Alberto Grimoldi A, Schnyder H. 2006. Phosphorus deficiency decreases cell division and elongation in grass leaves. Plant Physiol, 141(2): 766-775. |
| [50] | Kiba T, Inaba J, Kudo T, Ueda N, Konishi M, Mitsuda N, Takiguchi Y, Kondou Y, Yoshizumi T, Ohme-Takagi M, Matsui M, Yano K, Yanagisawa S, Sakakibara H. 2018. Repression of nitrogen starvation responses by members of the Arabidopsis GARP-type transcription factor NIGT1/HRS1 subfamily. Plant Cell, 30(4): 925-945. |
| [51] | Kumar S, Pallavi, Chugh C, Seem K, Kumar S, Vinod K K, Mohapatra T. 2021a. Characterization of contrasting rice (Oryza sativa L.) genotypes reveals the Pi-efficient schema for phosphate starvation tolerance. BMC Plant Biol, 21(1): 282. |
| [52] | Kumar S, Kumar S, Mohapatra T. 2021b. Interaction between macro- and micro-nutrients in plants. Front Plant Sci, 12: 665583. |
| [53] | Kumar S, Agrawal A, Seem K, Kumar S, Vinod K K, Mohapatra T. 2022a. Transcriptome analysis of a near-isogenic line and its recurrent parent reveals the role of Pup1 QTL in phosphorus deficiency tolerance of rice at tillering stage. Plant Mol Biol, 109(1/2): 29-50. |
| [54] | Kumar S, Seem K, Kumar S, Vinod K K, Chinnusamy V, Mohapatra T. 2022b. Pup1 QTL regulates gene expression through epigenetic modification of DNA under phosphate starvation stress in rice. Front Plant Sci, 13: 871890. |
| [55] | Lambers H, Brundrett M C, Raven J A, Hopper S D. 2011. Plant mineral nutrition in ancient landscapes: High plant species diversity on infertile soils is linked to functional diversity for nutritional strategies. Plant Soil, 348(1): 7-27. |
| [56] | Lauer M J, Pallardy S G, Blevins D G, Randall D D. 1989. Whole leaf carbon exchange characteristics of phosphate deficient soybeans (Glycine max L.). Plant Physiol, 91(3): 848-854. |
| [57] | Lee J Y, Colinas J, Wang J Y, Mace D, Ohler U, Benfey P N. 2006. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA, 103(15): 6055-6060. |
| [58] | Lee S K, Hwang S K, Han M, Eom J S, Kang H G, Han Y, Choi S B, Cho M H, Bhoo S H, An G, Hahn T R, Okita T W, Jeon J S. 2007. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol Biol, 65(4): 531-546. |
| [59] | Lei L, Li Y, Wang Q, Xu J, Chen Y F, Yang H L, Ren D T. 2014. Activation of MKK 9-MPK 3/MPK 6 enhances phosphate acquisition in Arabidopsis thaliana. New Phytol, 203(4): 1146-1160. |
| [60] | Lei M G, Liu Y D, Zhang B C, Zhao Y T, Wang X J, Zhou Y H, Raghothama K G, Liu D. 2011. Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol, 156(3): 1116-1130. |
| [61] | Li K P, Xu C Z, Li Z X, Zhang K W, Yang A F, Zhang J R. 2008. Comparative proteome analyses of phosphorus responses in maize (Zea mays L.) roots of wild-type and a low-P-tolerant mutant reveal root characteristics associated with phosphorus efficiency. Plant J, 55(6): 927-939. |
| [62] | Li M Y, Welti R, Wang X M. 2006. Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases Dζ1 and Dζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol, 142(2): 750-761. |
| [63] | Li Z Y, Hu J Y, Wu Y, Wang J X, Song H, Chai M F, Cong L L, Miao F H, Ma L C, Tang W, Yang C, Tao Q B, Zhong S Z, Zhao Y R, Liu H Q, Yang G F, Wang Z Y, Sun J. 2022. Integrative analysis of the metabolome and transcriptome reveal the phosphate deficiency response pathways of alfalfa. Plant Physiol Biochem, 170: 49-63. |
| [64] | Liang C Y, Wang J X, Zhao J, Tian J, Liao H. 2014. Control of phosphate homeostasis through gene regulation in crops. Curr Opin Plant Biol, 21: 59-66. |
| [65] | Liang G, Ai Q, Yu D Q. 2015. Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci Rep, 5: 11813. |
| [66] | Lin W Y, Huang T K, Chiou T J. 2013. Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell, 25(10): 4061-4074. |
| [67] | Liu H J, Wang J C, Zhang B B, Yang X Y, Hammond J P, Ding G D, Wang S L, Cai H M, Wang C, Xu F S, Shi L. 2021. Genome-wide association study dissects the genetic control of plant height and branch number in response to low-phosphorus stress in Brassica napus. Ann Bot, 128(7): 919-930. |
| [68] | Liu J Q, Vance C P. 2010. Crucial roles of sucrose and microRNA399 in systemic signaling of P deficiency: A tale of two team players? Plant Signal Behav, 5(12): 1556-1560. |
| [69] | López-Arredondo D L, Leyva-González M A, González-Morales S I, López-Bucio J, Herrera-Estrella L. 2014. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu Rev Plant Biol, 65: 95-123. |
| [70] | Lutfiyya L L, Xu N F, D’Ordine R L, Morrell J A, Miller P W, Duff S M G. 2007. Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J Plant Physiol, 164(7): 923-933. |
| [71] | Lynch J P. 2011. Root Phenes for enhanced soil exploration and phosphorus acquisition: Tools for future crops. Plant Physiol, 156(3): 1041-1049. |
| [72] | Ma Z, Baskin T I, Brown K M, Lynch J P. 2003. Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol, 131(3): 1381-1390. |
| [73] | Maeda Y, Konishi M, Kiba T, Sakuraba Y, Sawaki N, Kurai T, Ueda Y, Sakakibara H, Yanagisawa S. 2018. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat Commun, 9: 1376. |
| [74] | Medici A, Szponarski W, Dangeville P, Safi A, Dissanayake I M, Saenchai C, Emanuel A, Rubio V, Lacombe B, Ruffel S, Tanurdzic M, Rouached H, Krouk G. 2019. Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell, 31(5): 1171-1184. |
| [75] | Meng Q, Zhang W Q, Hu X, Shi X Y, Chen L L, Dai X L, Qu H Y, Xia Y W, Liu W, Gu M, Xu G H. 2020. Two ADP-glucose pyrophosphorylase subunits, OsAGPL1 and OsAGPS1, modulate phosphorus homeostasis in rice. Plant J, 104(5): 1269-1284. |
| [76] | Mghase J J, Shiwachi H, Takahashi H, Irie K. 2011. Nutrient deficiencies and their symptoms in upland rice. J Int Soc Southeast Asian Agric Sci, 17: 59-67. |
| [77] | Morcuende R, Bari R, Gibon Y, Zheng W M, Datt Pant B, Bläsing O, Usadel B, Czechowski T, Udvardi M K, Stitt M, Scheible W R. 2007. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ, 30(1): 85-112. |
| [78] | Müller R, Morant M, Jarmer H, Nilsson L, Nielsen T H. 2007. Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol, 143(1): 156-171. |
| [79] | Muralidharudu Y, Reddy, K Sammi, Mandal B N, Subba Rao A, Singh K N, Sonekar S. 2011. GIS based soil fertility maps of different states of India. In: All India Coordinated Project on Soil Test Crop Response Correlation. Bhopal: Indian Institute of Soil Science: 224. |
| [80] | Nanamori M, Shinano T, Wasaki J, Yamamura T, Rao I M, Osaki M. 2004. Low phosphorus tolerance mechanisms: Phosphorus recycling and photosynthate partitioning in the tropical forage grass, Brachiaria hybrid cultivar mulato compared with rice. Plant Cell Physiol, 45(4): 460-469. |
| [81] | Narang R A, Altmann T. 2001. Phosphate acquisition heterosis in Arabidopsis thaliana: A morphological and physiological analysis. Plant Soil, 234(1): 91-97. |
| [82] | Nguyen V N T, Lee S B, Suh M C, An G, Jung K H. 2018. OsABCG9 is an important ABC transporter of cuticular wax deposition in rice. Front Plant Sci, 9: 960. |
| [83] | Ni J J, Wu P, Senadhira D, Huang N. 1998. Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Theor Appl Genet, 97(8): 1361-1369. |
| [84] | Nielsen T H, Krapp A, Röper-Schwarz U, Stitt M. 1998. The sugar-mediated regulation of genes encoding the small subunit of Rubisco and the regulatory subunit of ADP glucose pyrophosphorylase is modified by phosphate and nitrogen. Plant Cell Environ, 21(5): 443-454. |
| [85] | Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi T M, Thibaud M C. 2011. Phosphate import in plants: Focus on the PHT1 transporters. Front Plant Sci, 2: 83. |
| [86] | Pariasca-Tanaka J, Satoh K, Rose T, Mauleon R, Wissuwa M. 2009. Stress response versus stress tolerance: A transcriptome analysis of two rice lines contrasting in tolerance to phosphorus deficiency. Rice, 2(4): 167-185. |
| [87] | Peng M S, Hannam C, Gu H L, Bi Y M, Rothstein S J. 2007. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J, 50(2): 320-337. |
| [88] | Péret B, Clément M, Nussaume L, Desnos T. 2011. Root developmental adaptation to phosphate starvation: Better safe than sorry. Trends Plant Sci, 16(8): 442-450. |
| [89] | Pieters A J, Paul M J, Lawlor D W. 2001. Low sink demand limits photosynthesis under Pi deficiency. J Exp Bot, 52(358): 1083-1091. |
| [90] | Plaxton W C, Tran H T. 2011. Metabolic adaptations of phosphate- starved plants. Plant Physiol, 156(3): 1006-1015. |
| [91] | Plaxton W, Lambers H. 2015. Annual Plant Reviews: Phosphorus Metabolism in Plants. Manhattan, USA: John Wiley & Sons, Ltd.: 48. |
| [92] | Prathap V, Kumar A, Maheshwari C, Tyagi A. 2022. Phosphorus homeostasis: Acquisition, sensing, and long-distance signaling in plants. Mol Biol Rep, 49(8): 8071-8086. |
| [93] | Qi Q T, Hong Y, Zhang Y Y, Gu Z B, Cheng L, Li Z F, Li C M. 2020. Combinatorial effect of fermentation and drying on the relationship between the structure and expansion properties of tapioca starch and potato starch. Int J Biol Macromol, 145: 965-973. |
| [94] | Raghothama K G. 1999. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol, 50: 665-693. |
| [95] | Rao I M, Fredeen A L, Terry N. 1990. Leaf phosphate status, photosynthesis, and carbon partitioning in sugar beet: III. Diurnal changes in carbon partitioning and carbon export. Plant Physiol, 92(1): 29-36. |
| [96] | Rausch C, Bucher M. 2002. Molecular mechanisms of phosphate transport in plants. Planta, 216(1): 23-37. |
| [97] | Rengel Z, Marschner P. 2005. Nutrient availability and management in the rhizosphere: Exploiting genotypic differences. New Phytol, 168(2): 305-312. |
| [98] | Ruan W Y, Guo M N, Xu L, Wang X Q, Zhao H Y, Wang J M, Yi K K. 2018. An SPX-RLI1 module regulates leaf inclination in response to phosphate availability in rice. Plant Cell, 30(4): 853-870. |
| [99] | Rubio V, Linhares F, Solano R, Martín A C, Iglesias J, Leyva A, Paz-Ares J. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev, 15(16): 2122-2133. |
| [100] | Sano H, Youssefian S. 1994. Light and nutritional regulation of transcripts encoding a wheat protein kinase homolog is mediated by cytokinins. Proc Natl Acad Sci USA, 91(7): 2582-2586. |
| [101] | Scheible W R, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi M K, Stitt M. 2004. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol, 136(1): 2483-2499. |
| [102] | Scheible W R, Rojas-Triana M. 2015. Sensing, signalling, and control of phosphate starvation in plants: Molecular players and applications. Annu Plant Rev, 48: 23-63. |
| [103] | Secco D, Wang C, Shou H X, Schultz M D, Chiarenza S, Nussaume L, Ecker J R, Whelan J, Lister R. 2015. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife, 4: e09343. |
| [104] | Shabnam R, Iqbal M T. 2016. Understanding phosphorus dynamics on wheat plant under split-root system in alkaline soil. Braz J Sci Technol, 3(1): 1-16. |
| [105] | Sharma S, Borah P, Meena M K, Bindraban P, Pandey R. 2018. Evaluation of genotypic variation for growth of rice seedlings under optimized hydroponics medium. Indian J Genet Pl Br, 78: 292-301. |
| [106] | Sharma V, Thaker H. 2011. Demand for fertilizer in India: Determinants and outlook for 2020. India: Indian Institute of Management Ahmedabad. |
| [107] | Shimizu A, Kato K, Komatsu A, Motomura K, Ikehashi H. 2008. Genetic analysis of root elongation induced by phosphorus deficiency in rice (Oryza sativa L.): Fine QTL mapping and multivariate analysis of related traits. Theor Appl Genet, 117(6): 987-996. |
| [108] | Shobbar Z S, Bennett J. 2012. Identification and functional analysis of an ABA and drought stress inducible protein phosphatase 2C in rice. Crop Biotechnol, 1(1): 23-33. |
| [109] | Siebers M, Dörmann P, Hölzl G. 2015. Membrane remodelling in phosphorus-deficient plants. Annu Plant Rev, 48: 237-263. |
| [110] | Stigter K A, Plaxton W C. 2015. Molecular mechanisms of phosphorus metabolism and transport during leaf senescence. Plants, 4(4): 773-798. |
| [111] | Stitt M, Quick W P. 1989. Photosynthetic carbon partitioning: Its regulation and possibilities for manipulation. Physiol Plant, 77(4): 633-641. |
| [112] | Sun Y F, Luo W Z, Jain A, Liu L, Ai H, Liu X L, Feng B, Zhang L, Zhang Z T, Xu G H, Sun S B. 2018. OsPHR3 affects the traits governing nitrogen homeostasis in rice. BMC Plant Biol, 18(1): 241. |
| [113] | Sun Y L, Mu C H, Chen Y, Kong X P, Xu Y C, Zheng H X, Zhang H, Wang Q C, Xue Y F, Li Z X, Ding Z J, Liu X. 2016. Comparative transcript profiling of maize inbreds in response to long-term phosphorus deficiency stress. Plant Physiol Biochem, 109: 467-481. |
| [114] | Tantray A Y, Ali H M, Ahmad A. 2020. Analysis of proteomic profile of contrasting phosphorus responsive rice cultivars grown under phosphorus deficiency. Agronomy, 10(7): 1028. |
| [115] | Tian J, Wang X R, Tong Y P, Chen X P, Liao H. 2012. Bioengineering and management for efficient phosphorus utilization in crops and pastures. Curr Opin Biotechnol, 23(6): 866-871. |
| [116] | Tran H T, Hurley B A, Plaxton W C. 2010. Feeding hungry plants: The role of purple acid phosphatases in phosphate nutrition. Plant Sci, 179(1/2): 14-27. |
| [117] | Tsai T H, Wang M K, Ressom H W. 2016. Preprocessing and analysis of LC-MS-based proteomic data. Methods Mol Biol, 1362: 63-76. |
| [118] | Vance C P, Uhde-Stone C, Allan D L. 2003. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol, 157(3): 423-447. |
| [119] | Varala K, Marshall-Colón A, Cirrone J, Brooks M D, Pasquino A V, Léran S, Mittal S, Rock T M, Edwards M B, Kim G J, Ruffel S, Richard McCombie W, Shasha D, Coruzzi G M. 2018. Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc Natl Acad Sci USA, 115(25): 6494-6499. |
| [120] | Vinod K K, Heuer S. 2012. Approaches towards nitrogen- and phosphorus-efficient rice. AoB PLANTS, 2012: pls028. |
| [121] | Wang Q, Du W K, Yu W Q, Zhang W H, Huang F, Cheng H, Yu D Y. 2022. Genome-wide association analysis discovered new loci and candidate genes associated with low-phosphorus tolerance based on shoot mineral elements concentrations in soybean. Mol Genet Genom, 297(3): 843-858. |
| [122] | Wang W N, Wang A L, Chen L, Liu Y, Sun R Y. 2002. Effects of pH on survival, phosphorus concentration, adenylate energy charge and Na+-K+ ATPase activities of Penaeus chinensis Osbeck juveniles. Aquat Toxicol, 60(1/2): 75-83. |
| [123] | Wang X R, Shen J B, Liao H. 2010. Acquisition or utilization, which is more critical for enhancing phosphorus efficiency in modern crops? Plant Sci, 179(4): 302-306. |
| [124] | Wang Y L, Lysøe E, Armarego-Marriott T, Erban A, Paruch L, van Eerde A, Bock R, Liu-Clarke J. 2018. Transcriptome and metabolome analyses provide insights into root and root-released organic anion responses to phosphorus deficiency in oat. J Exp Bot, 69(15): 3759-3771. |
| [125] | White P J, Hammond J P. 2008. Phosphorus nutrition of terrestrial plants. In: White P J, Hammond J P. The Ecophysiology of Plant-Phosphorus Interactions: Plant Ecophysiology. Dordrecht, the Netherlands: Springer: 51-81. |
| [126] | Wissuwa M, Ae N. 2001. Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breed, 120(1): 43-48. |
| [127] | Wissuwa M, Yano M, Ae N. 1998. Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa L.). Theor Appl Genet, 97: 777-783. |
| [128] | Wissuwa M, Wegner J, Ae N, Yano M. 2002. Substitution mapping of Pup1: A major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor Appl Genet, 105: 890-897. |
| [129] | Wissuwa M, Gamat G, Ismail A M. 2005. Is root growth under phosphorus deficiency affected by source or sink limitations? J Exp Bot, 56: 1943-1950. |
| [130] | Xing D, Wu Y. 2014. Effect of phosphorus deficiency on photosynthetic inorganic carbon assimilation of three climber plant species. Bot Stud, 55(1): 1-8. |
| [131] | Xu H X, Weng X Y, Yang Y. 2007. Effect of phosphorus deficiency on the photosynthetic characteristics of rice plants. Russ J Plant Physiol, 54: 741-748. |
| [132] | Xue T T, Wang D, Zhang S Z, Ehlting J, Ni F, Jakab S, Zheng C C, Zhong Y. 2008. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genom, 9(1): 1-21. |
| [133] | Yang J, Xie M Y, Yang X L, Liu B H, Lin H H. 2019. Phosphoproteomic profiling reveals the importance of CK2, MAPKs and CDPKs in response to phosphate starvation in rice. Plant Cell Physiol, 60(12): 2785-2796. |
| [134] | Yang Z L, Yang J, Wang Y, Wang F, Mao W X, He Q J, Xu J M, Wu Z C, Mao C Z. 2020. PROTEIN PHOSPHATASE95 regulates phosphate homeostasis by affecting phosphate transporter trafficking in rice. Plant Cell, 32(3): 740-757. |
| [135] | Yao Y N, Sun H Y, Xu F S, Zhang X J, Liu S Y. 2011. Comparative proteome analysis of metabolic changes by low phosphorus stress in two Brassica napus genotypes. Planta, 233(3): 523-537. |
| [136] | Yau C P, Wang L J, Yu M D, Zee S Y, Yip W K. 2004. Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions. J Exp Bot, 55(397): 547-556. |
| [137] | Yuan H, Liu D. 2008. Signaling components involved in plant responses to phosphate starvation. J Integr Plant Biol, 50(7): 849-859. |
| [138] | Yugandhar P, Veronica N, Panigrahy M, Nageswara Rao D, Subrahmanyam D, Voleti S R, Mangrauthia S K, Sharma R P, Sarla N. 2017. Comparing hydroponics, sand, and soil medium to evaluate contrasting rice nagina 22 mutants for tolerance to phosphorus deficiency. Crop Sci, 57(4): 2089-2097. |
| [139] | Zhang H J, Huang L, Hong Y B, Song F M. 2016. BOTRYTIS- INDUCED KINASE1, a plasma membrane-localized receptor- like protein kinase, is a negative regulator of phosphate homeostasis in Arabidopsis thaliana. BMC Plant Biol, 16(1): 152. |
| [140] | Zhang J, Xu H F, Wang N, Jiang S H, Fang H C, Zhang Z Y, Yang G X, Wang Y C, Su M Y, Xu L, Chen X S. 2018. The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol Biol, 98(3): 205-218. |
| [141] | Zhang J L, Jiang F F, Shen Y X, Zhan Q W, Bai B Q, Chen W, Chi Y J. 2019. Transcriptome analysis reveals candidate genes related to phosphorus starvation tolerance in sorghum. BMC Plant Biol, 19(1): 306. |
| [142] | Zhang K W, Liu H H, Tao P L, Chen H. 2014. Comparative proteomic analyses provide new insights into low phosphorus stress responses in maize leaves. PLoS One, 9(5): e98215. |
| [143] | Zhang Q, Wang C, Tian J, Li K, Shou H. 2011. Identification of rice purple acid phosphatases related to posphate starvation signalling. Plant Biol, 13(1): 7-15. |
| [144] | Zhao B B, Sun S W, Lin H, Chen L D, Qin S, Wu W G, Zheng B D, Guo Z B. 2019. Physicochemical properties and digestion of the lotus seed starch-green tea polyphenol complex under ultrasound-microwave synergistic interaction. Ultrason Sonochem, 52: 50-61. |
| [145] | Zhong Y J, Wang Y G, Guo J F, Zhu X L, Shi J, He Q J, Liu Y, Wu Y R, Zhang L, Lv Q D, Mao C Z. 2018. Rice SPX6 negatively regulates the phosphate starvation response through suppression of the transcription factor PHR2. New Phytol, 219(1): 135-148. |
| [146] | Zhou J, Jiao F C, Wu Z C, Li Y Y, Wang X M, He X W, Zhong W Q, Wu P. 2008. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol, 146(4): 1673-1686. |
| [1] | Li Qianlong, Feng Qi, Wang Heqin, Kang Yunhai, Zhang Conghe, Du Ming, Zhang Yunhu, Wang Hui, Chen Jinjie, Han Bin, Fang Yu, Wang Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 552-565. |
| [2] | Ji Dongling, Xiao Wenhui, Sun Zhiwei, Liu Lijun, Gu Junfei, Zhang Hao, Matthew Tom Harrison, Liu Ke, Wang Zhiqin, Wang Weilu. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 598-612. |
| [3] | Serena Reggi, Elisabetta Onelli, Alessandra Moscatelli, Nadia Stroppa, Matteo Dell’Anno, Kiril Perfanov, Luciana Rossi. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Engineered Rice Lines [J]. Rice Science, 2023, 30(6): 587-597. |
| [4] | Sundus Zafar, Xu Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 523-536. |
| [5] | Kankunlanach Khampuang, Nanthana Chaiwong, Atilla Yazici, Baris Demirer, Ismail Cakmak, Chanakan Prom-U-Thai. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 632-640. |
| [6] | Fan Fengfeng, Cai Meng, Luo Xiong, Liu Manman, Yuan Huanran, Cheng Mingxing, Ayaz Ahmad, Li Nengwu, Li Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 577-586. |
| [7] | Lin Shaodan, Yao Yue, Li Jiayi, Li Xiaobin, Ma Jie, Weng Haiyong, Cheng Zuxin, Ye Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 652-660. |
| [8] | Md. Forshed Dewan, Md. Ahiduzzaman, Md. Nahidul Islam, Habibul Bari Shozib. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and Southeast Asia: A Review [J]. Rice Science, 2023, 30(6): 537-551. |
| [9] | Raja Chakraborty, Pratap Kalita, Saikat Sen. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Pigmented Black Rice Variety Chakhao poireiton in High-Fat High-Sugar Induced Rats [J]. Rice Science, 2023, 30(6): 641-651. |
| [10] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [11] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [12] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [13] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [14] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| [15] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||