
Rice Science ›› 2025, Vol. 32 ›› Issue (1): 32-43.DOI: 10.1016/j.rsci.2024.12.003
• Reviews • Previous Articles Next Articles
Hao Zhiqi1,#, Wang Tingyi1,#, Chen Dongdong1, Shen Lan1, Zhang Guangheng1,3, Qian Qian1,2, Zhu Li1,3( )
)
Received:2024-07-29
Accepted:2024-12-10
Online:2025-01-28
Published:2025-03-25
Contact:
Zhu Li
About author:First author contact:#These authors contributed equally to this work
Hao Zhiqi, Wang Tingyi, Chen Dongdong, Shen Lan, Zhang Guangheng, Qian Qian, Zhu Li. Leucine-Rich Repeat Protein Family Regulates Stress Tolerance and Development in Plants[J]. Rice Science, 2025, 32(1): 32-43.
Add to citation manager EndNote|Ris|BibTeX
| Type | Gene | Function | LRR location | No. of LRRs | Plant origin | Reference |
|---|---|---|---|---|---|---|
| LRR-RLK | ERECTA | Transpiration efficiency | N-terminus | 18 | Arabidopsis thaliana | Masle et al, |
| OsSIK1 | Drought and salt stress | N-terminus | 16 | Oryza sativa | Ouyang et al, | |
| FLS2 | Signaling activation; Mediated immunity | N-terminus | 25 | A. thaliana | Cao et al, | |
| PXC1 | Secondary cell wall formation; Tracheary element differentiation inhibitory factor-phloem intercalated with xylem/TDIF receptor-WOX4 signaling | N-terminus | 4 | A. thaliana | Wang J H et al, | |
| LIK1 | Immune responses | N-terminus | 10 | A. thaliana | Le et al, | |
| OsGIRL1 | Abiotic stress | N-terminus | - | O. sativa | Park et al, | |
| NIK1 | Geminivirus resistance | N-terminus | 4 | A. thaliana | Zorzatto et al, | |
| OsWAK25 | Biological stress | N-terminus | - | O. sativa | Harkenrider et al, | |
| MRK1 | Temperature stress; Resistance to bacterial disease | N-terminus | 5 | Solanum lycopersicum | Ma et al, | |
| RINRK1 | Nodulation factor signaling; Rhizobia infection | N-terminus | 3 | Lotus japonicus | Li X L et al, | |
| ZmRLK7 | Enlarges organ and seed size | N-terminus | 10 | Zea mays | He et al, | |
| HSL3 | Drought stress | N-terminus | 18 | A. thaliana | Liu et al, | |
| MtCTLK1 | Cold tolerance | N-terminus | 3 | Medicago truncatula | Geng et al, | |
| OsCERK1 | Chitin signaling | N-terminus | - | O. sativa | Yang et al, | |
| OsBAK1 | Immune regulation | N-terminus | 4 | O. sativa | Duan et al, | |
| LRR-RLP | HcrVf2 | Resistance to Venturia inaequalis | N-terminus | 27 | Malus floribunda | Belfanti et al, |
| ReMAX | Perception of enigmatic MAMP protein of Xanthomonas | N-terminus | 27 | A. thaliana | Jehle et al, | |
| RFO2 | Resistance to Fusarium oxysporum | N-terminus | 16 | A. thaliana | Shen and Diener, | |
| VE1 | Resistance to Verticillium dahliae and V. albo-atrum | N-terminus | 30 | S. lycopersicum | Nazar et al, | |
| OsRLP1 | Resistance to rice black-streaked dwarf virus | N-terminus | 26 | O. sativa | Zhang H H et al, | |
| COG1 | Cold stress response | N-terminus | 17 | O. sativa | Xia et al, | |
| NBS-LRR | RPM1 | Resistance to Pseudomonas syringae | C-terminus | 9 | A. thaliana | Tornero et al, |
| Rxo1/Rba1 | Non-host resistance and resistance to pathogens | C-terminus | 9 | Z. mays | Zhao et al, | |
| BNT1 | Alters plant stress hormone levels | C-terminus | 11 | A. thaliana | Sarazin et al, | |
| Pm21 | Confers powdery mildew resistance | C-terminus | 6 | Haynaldia villosa | Xing et al, | |
| OsPi304 | Cold stress response | C-terminus | - | O. sativa | Yang et al, | |
| GhDSC1 | Resistance to Verticillium wilt | C-terminus | 4 | Gossypium hirsutum | Li T G et al, | |
| Roq1 | Interacts with Xanthomonas outer protein Q and triggers hypersensitive cell death | C-terminus | 24 | Nicotiana benthamiana | Martin et al, | |
| L5 | Induces cell death and self-association | C-terminus | 2 | A. thaliana | Huang et al, | |
| SNC1 | Immune activation | C-terminus | 19 | A. thaliana | Jia et al, | |
| MdTNL1 | Resistance to Glomerella leaf spot | C-terminus | 7 | M. domestica | Lv et al, | |
| RppM | Resistance to Southern Corn Rust | C-terminus | 17 | Z. mays | Wang S et al, | |
| LRX | LRX1 | Cell wall formation | N-terminus | 8 | A. thaliana | Draeger et al, |
| LRX2 | Cell wall formation | N-terminus | 8 | A. thaliana | Draeger et al, | |
| LRX3 | Regulates growth and salt tolerance | N-terminus | 8 | A. thaliana | Zhao et al, | |
| LRX4 | Regulates growth and salt tolerance | N-terminus | 8 | A. thaliana | Zhao et al, | |
| LRX5 | Regulates growth and salt tolerance | N-terminus | 8 | A. thaliana | Zhao et al, | |
| PGIP | AtPGIP1 | Resistance to Botrytis cinerea | C-terminus | 7 | A. thaliana | Ferrari et al, |
| GmPGIP1 | Resistance to Fusarium moniliforme | C-terminus | 9 | Glycine max | Maulik et al, | |
| PvPGIP1 | Resistance to F. moniliforme | C-terminus | 8 | Phaseolus vulgaris | Maulik et al, | |
| OsPGIP2 | Polygalacturonase inhibition | C-terminus | 6 | O. sativa | Chen et al, | |
| OsPGIP1 | Bacterial leaf streak tolerance | C-terminus | 7 | O. sativa | Wu et al, | |
| OsPGIP4 | Bacterial leaf streak | C-terminus | 8 | O. sativa | Wu et al, | |
| VrPGIP2 | Resistance to bruchids | C-terminus | 8 | Vigna radiata | Zhang Q et al, |
Table 1. Selected members of LRR protein family in plants.
| Type | Gene | Function | LRR location | No. of LRRs | Plant origin | Reference |
|---|---|---|---|---|---|---|
| LRR-RLK | ERECTA | Transpiration efficiency | N-terminus | 18 | Arabidopsis thaliana | Masle et al, |
| OsSIK1 | Drought and salt stress | N-terminus | 16 | Oryza sativa | Ouyang et al, | |
| FLS2 | Signaling activation; Mediated immunity | N-terminus | 25 | A. thaliana | Cao et al, | |
| PXC1 | Secondary cell wall formation; Tracheary element differentiation inhibitory factor-phloem intercalated with xylem/TDIF receptor-WOX4 signaling | N-terminus | 4 | A. thaliana | Wang J H et al, | |
| LIK1 | Immune responses | N-terminus | 10 | A. thaliana | Le et al, | |
| OsGIRL1 | Abiotic stress | N-terminus | - | O. sativa | Park et al, | |
| NIK1 | Geminivirus resistance | N-terminus | 4 | A. thaliana | Zorzatto et al, | |
| OsWAK25 | Biological stress | N-terminus | - | O. sativa | Harkenrider et al, | |
| MRK1 | Temperature stress; Resistance to bacterial disease | N-terminus | 5 | Solanum lycopersicum | Ma et al, | |
| RINRK1 | Nodulation factor signaling; Rhizobia infection | N-terminus | 3 | Lotus japonicus | Li X L et al, | |
| ZmRLK7 | Enlarges organ and seed size | N-terminus | 10 | Zea mays | He et al, | |
| HSL3 | Drought stress | N-terminus | 18 | A. thaliana | Liu et al, | |
| MtCTLK1 | Cold tolerance | N-terminus | 3 | Medicago truncatula | Geng et al, | |
| OsCERK1 | Chitin signaling | N-terminus | - | O. sativa | Yang et al, | |
| OsBAK1 | Immune regulation | N-terminus | 4 | O. sativa | Duan et al, | |
| LRR-RLP | HcrVf2 | Resistance to Venturia inaequalis | N-terminus | 27 | Malus floribunda | Belfanti et al, |
| ReMAX | Perception of enigmatic MAMP protein of Xanthomonas | N-terminus | 27 | A. thaliana | Jehle et al, | |
| RFO2 | Resistance to Fusarium oxysporum | N-terminus | 16 | A. thaliana | Shen and Diener, | |
| VE1 | Resistance to Verticillium dahliae and V. albo-atrum | N-terminus | 30 | S. lycopersicum | Nazar et al, | |
| OsRLP1 | Resistance to rice black-streaked dwarf virus | N-terminus | 26 | O. sativa | Zhang H H et al, | |
| COG1 | Cold stress response | N-terminus | 17 | O. sativa | Xia et al, | |
| NBS-LRR | RPM1 | Resistance to Pseudomonas syringae | C-terminus | 9 | A. thaliana | Tornero et al, |
| Rxo1/Rba1 | Non-host resistance and resistance to pathogens | C-terminus | 9 | Z. mays | Zhao et al, | |
| BNT1 | Alters plant stress hormone levels | C-terminus | 11 | A. thaliana | Sarazin et al, | |
| Pm21 | Confers powdery mildew resistance | C-terminus | 6 | Haynaldia villosa | Xing et al, | |
| OsPi304 | Cold stress response | C-terminus | - | O. sativa | Yang et al, | |
| GhDSC1 | Resistance to Verticillium wilt | C-terminus | 4 | Gossypium hirsutum | Li T G et al, | |
| Roq1 | Interacts with Xanthomonas outer protein Q and triggers hypersensitive cell death | C-terminus | 24 | Nicotiana benthamiana | Martin et al, | |
| L5 | Induces cell death and self-association | C-terminus | 2 | A. thaliana | Huang et al, | |
| SNC1 | Immune activation | C-terminus | 19 | A. thaliana | Jia et al, | |
| MdTNL1 | Resistance to Glomerella leaf spot | C-terminus | 7 | M. domestica | Lv et al, | |
| RppM | Resistance to Southern Corn Rust | C-terminus | 17 | Z. mays | Wang S et al, | |
| LRX | LRX1 | Cell wall formation | N-terminus | 8 | A. thaliana | Draeger et al, |
| LRX2 | Cell wall formation | N-terminus | 8 | A. thaliana | Draeger et al, | |
| LRX3 | Regulates growth and salt tolerance | N-terminus | 8 | A. thaliana | Zhao et al, | |
| LRX4 | Regulates growth and salt tolerance | N-terminus | 8 | A. thaliana | Zhao et al, | |
| LRX5 | Regulates growth and salt tolerance | N-terminus | 8 | A. thaliana | Zhao et al, | |
| PGIP | AtPGIP1 | Resistance to Botrytis cinerea | C-terminus | 7 | A. thaliana | Ferrari et al, |
| GmPGIP1 | Resistance to Fusarium moniliforme | C-terminus | 9 | Glycine max | Maulik et al, | |
| PvPGIP1 | Resistance to F. moniliforme | C-terminus | 8 | Phaseolus vulgaris | Maulik et al, | |
| OsPGIP2 | Polygalacturonase inhibition | C-terminus | 6 | O. sativa | Chen et al, | |
| OsPGIP1 | Bacterial leaf streak tolerance | C-terminus | 7 | O. sativa | Wu et al, | |
| OsPGIP4 | Bacterial leaf streak | C-terminus | 8 | O. sativa | Wu et al, | |
| VrPGIP2 | Resistance to bruchids | C-terminus | 8 | Vigna radiata | Zhang Q et al, |
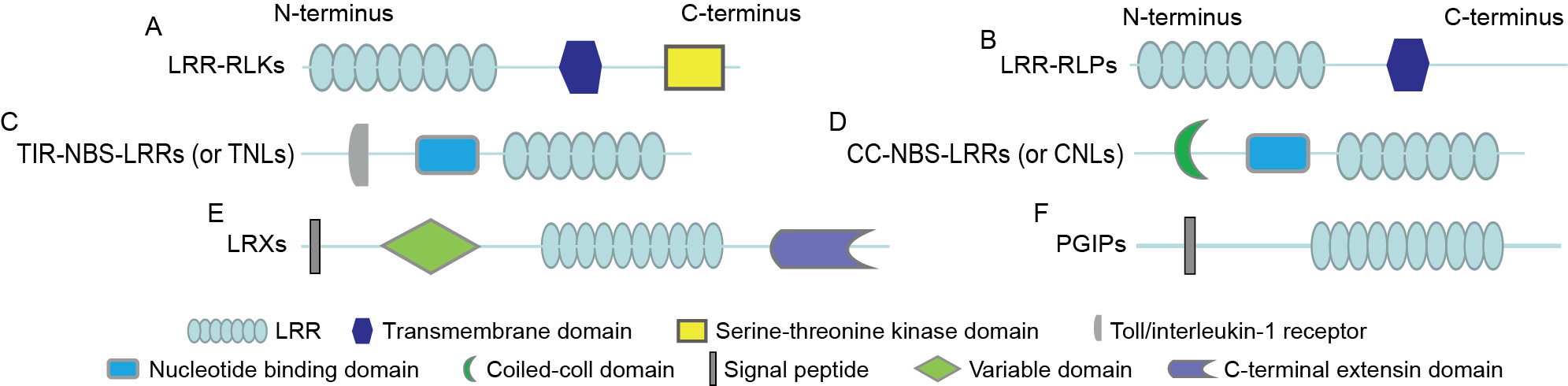
Fig. 1. Structures of leucine-rich repeat (LRR) receptor-like kinases (LRR-RLKs) and LRR receptor-like proteins (LRR-RLPs). A, Structural diagram of LRR-RLKs, which comprise an LRR domain, a transmembrane domain, and a kinase domain. B, Structural diagram of LRR-RLPs, which lack a kinase domain. C, Structure of Toll/interleukin-1 receptor (TIR) nucleotide-binding site LRR proteins (NBS-LRRs) (TIR-NBS-LRRs or TNLs). D, Structure of coiled-coil (CC) NBS-LRR (CC-NBS-LRRs or CNLs). E, Structure of LRR-extensin proteins (LRXs), which contain a signal peptide, a variable region, and an LRR domain, along with an extensin domain at the C-terminus. F, Schematic diagram of polygalacturonase-inhibiting proteins (PGIPs).
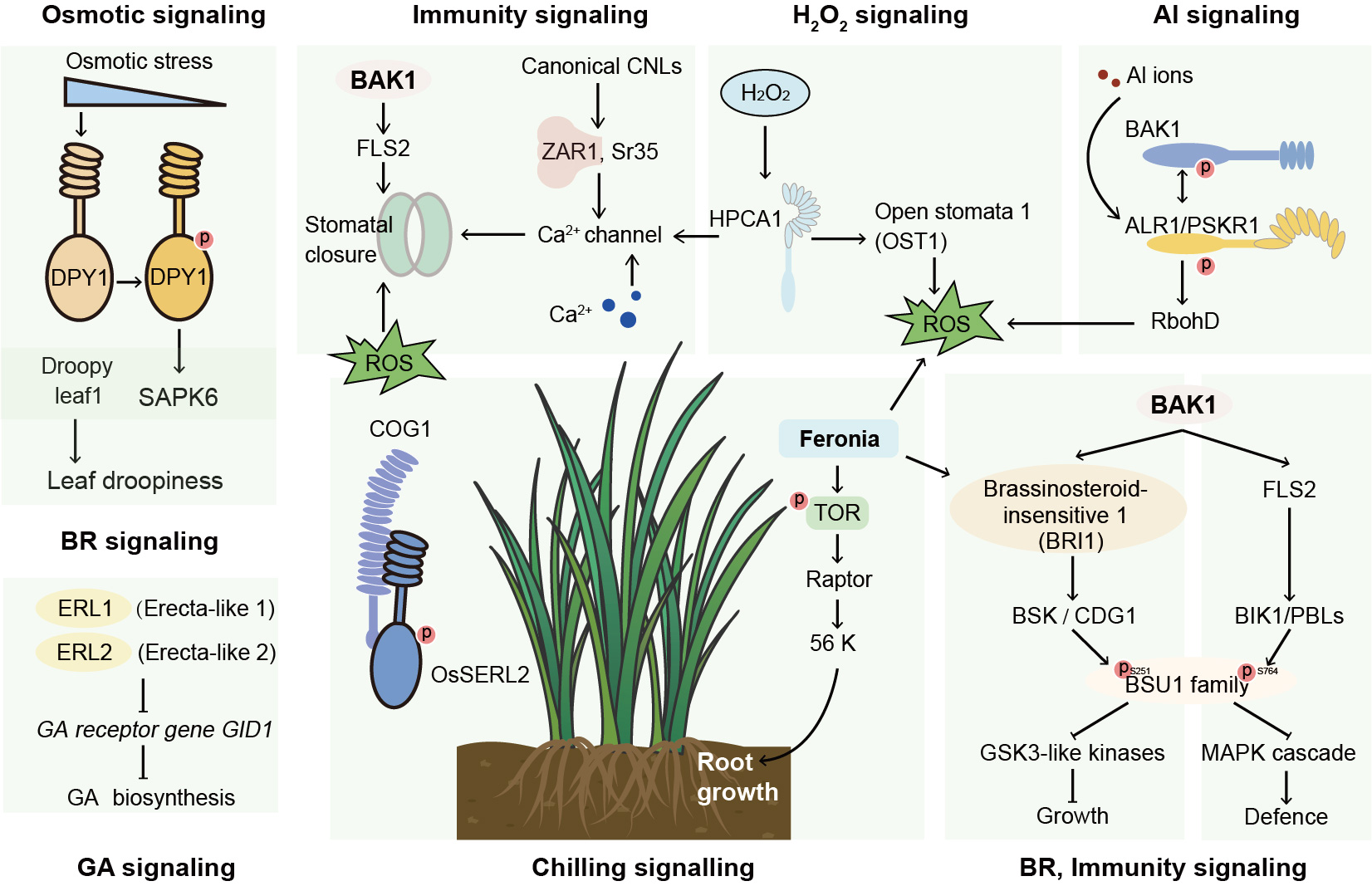
Fig. 2. Mechanisms of plant leucine-rich repeats (LRRs) in different stress signal transduction pathways. LRR family proteins are widely involved in various signal transduction processes in plants. Both the LRR receptor-like kinase (LRR-RLK) BAK1 and droopy leaf1 (DPY1) are involved in the brassinosteroid (BR) signaling pathway. DPY1 plays a crucial role in determining leaf droopiness by controlling BR signaling output. It is also important for sensing changes in osmotic potential and for intracellular signal transduction. Moreover, BAK1 has important function in the immune response. BAK1 interacts with FLS2 to enhance the generation of reactive oxygen species (ROS) and regulate stomatal closure. This interaction is crucial in the early defense of plants against bacteria. BAK1 also cooperates with other LRR-RLK receptors to regulate pathogen-associated defense responses. BRI1 and FLS2 phosphorylate different sites on bri1-suppressor1 (BSU1) phosphatase, which regulates the activity of downstream glycogen synthase kinase 3 (GSK3) and mitogen-activated protein kinase (MAPK) signaling pathways, respectively. The nucleotide-binding site and LRR receptor (NLR) protein ZAR1 and Sr35 resistosome can insert into lipid bilayer membranes to form cation channels with Ca2+ ion permeability, thereby performing immune response functions. The LRR-RLK hydrogen-peroxide-induced Ca2+ increase 1 (HPCA1) functions as a cellular H2O2 sensor that affects the activation of Ca2+ channels in guard cells via H2O2 signaling, the affecting the opening and closing of the stomata. In rice, the LRR-RLK ALR1 (aluminum resistance1) confers resistance to aluminum (Al) toxicity through an integrated Al-triggered signaling pathway. ALR1 binds Al ions, and its cytoplasmic domain recruits the BAK1 co-receptor kinase, promoting ALR1-dependent phosphorylation of RbohD by NADPH oxidase, thereby enhancing ROS production. The LRR-RLK ERECTA-LIKE 1 (ERL1) and ERECTA-LIKE 2 (ERL2) are involved in the gibberellin (GA) signaling pathway, affecting the expression of downstream GA reporter genes. The rice LRR-RLP Chilling-tolerance in Gengdao/japonica rice 1 (COG1) forms a complex with OsSERL2, further activate OsSERL2 perception of cold-stress signals in the plasma membrane. As a signaling hub, feronia (FER) regulates a variety of biotic and abiotic signaling pathways, enabling plants to respond to environmental changes and regulate growth and development. FER interacts with the target of rapamycin (TOR) protein in the TOR pathway, activating downstream components that trigger root hair growth under low-nutrient conditions. Additionally, FER cooperates with BRI1 to regulate plant cell growth, particularly during root and stem development.
| [1] | Ariza-Suarez D, Keller B, Spescha A, et al. 2023. Genetic analysis of resistance to bean leaf crumple virus identifies a candidate LRR-RLK gene. Plant J, 114(1): 23-38. |
| [2] | Baggs E, Dagdas G, Krasileva K V. 2017. NLR diversity, helpers and integrated domains: Making sense of the NLR IDentity. Curr Opin Plant Biol, 38: 59-67. |
| [3] | Belfanti E, Silfverberg-Dilworth E, Tartarini S, et al. 2004. The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc Natl Acad Sci USA, 101(3): 886-890. |
| [4] | Bella J, Hindle K L, McEwan P A, et al. 2008. The leucine-rich repeat structure. Cell Mol Life Sci, 65(15): 2307-2333. |
| [5] | Bi G Z, Su M, Li N, et al. 2021. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell, 184(13): 3528-3541.e12. |
| [6] | Cai H R, Wang W, Rui L, et al. 2021. The TIR-NBS protein TN13 associates with the CC-NBS-LRR resistance protein RPS5 and contributes to RPS5-triggered immunity in Arabidopsis. Plant J, 107(3): 775-786. |
| [7] | Cao Y, Aceti D J, Sabat G, et al. 2013. Mutations in FLS2 Ser-938 dissect signaling activation in FLS2-mediated Arabidopsis immunity. PLoS Pathog, 9(4): e1003313. |
| [8] | Cao Y P, Mo W Z, Li Y L, et al. 2021. Deciphering the roles of leucine-rich repeat receptor-like protein kinases (LRR-RLKs) in response to Fusarium wilt in the Vernicia fordii (Tung tree). Phytochemistry, 185: 112686. |
| [9] | Cao Y P, Mo W Z, Li Y L, et al. 2024. Functional characterization of NBS-LRR genes reveals an NBS-LRR gene that mediates resistance against Fusarium wilt. BMC Biol, 22(1): 45. |
| [10] | Castel B, El Mahboubi K, Jacquet C, et al. 2024. Immunobiodiversity: Conserved and specific immunity across land plants and beyond. Mol Plant, 17(1): 92-111. |
| [11] | Catanzariti A M, Do H T T, Bru P, et al. 2017. The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J, 89(6): 1195-1209. |
| [12] | Chen D D, Qiu Z N, He L, et al. 2021. The rice LRR-like1 protein YELLOW AND PREMATURE DWARF 1 is involved in leaf senescence induced by high light. J Exp Bot, 72(5): 1589-1605. |
| [13] | Chen J B, Bao S W, Fang Y L, et al. 2021. An LRR-only protein promotes NLP-triggered cell death and disease susceptibility by facilitating oligomerization of NLP in Arabidopsis. New Phytol, 232(4): 1808-1822. |
| [14] | Chen J P, Upadhyaya N M, Ortiz D, et al. 2017. Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science, 358: 1607-1610. |
| [15] | Chen X J, Chen Y W, Zhang L N, et al. 2019. Amino acid substitutions in a polygalacturonase inhibiting protein (OsPGIP2) increases sheath blight resistance in rice. Rice, 12(1): 56. |
| [16] | Cheung A Y. 2024. FERONIA: A receptor kinase at the core of a global signaling network. Annu Rev Plant Biol, 75(1): 345-375. |
| [17] | Chinchilla D, Shan L B, He P, et al. 2009. One for all: The receptor- associated kinase BAK1. Trends Plant Sci, 14(10): 535-541. |
| [18] | Colcombet J, Boisson-Dernier A, Ros-Palau R, et al. 2005. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell, 17(12): 3350-3361. |
| [19] | Covey P A, Subbaiah C C, Parsons R L, et al. 2010. A pollen- specific RALF from tomato that regulates pollen tube elongation. Plant Physiol, 153(2): 703-715. |
| [20] | da Silva Dambroz C M, Aono A H, de Andrade Silva E M, et al. 2023. Genome-wide analysis and characterization of the LRR- RLK gene family provides insights into anthracnose resistance in common bean. Sci Rep, 13(1): 13455. |
| [21] | de Lorenzo G, D’Ovidio R, Cervone F. 2001. The role of poly- galacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu Rev Phytopathol, 39: 313-335. |
| [22] | di Matteo A, Bonivento D, Tsernoglou D, et al. 2006. Poly- galacturonase-inhibiting protein (PGIP) in plant defence: A structural view. Phytochemistry, 67(6): 528-533. |
| [23] | Ding Z J, Xu C, Yan J Y, et al. 2024. The LRR receptor-like kinase ALR1 is a plant aluminum ion sensor. Cell Res, 34(4): 281-294. |
| [24] | Draeger C, Fabrice T N, Gineau E, et al. 2015. Arabidopsis leucine- rich repeat extensin (LRX) proteins modify cell wall composition and influence plant growth. BMC Plant Biol, 15: 155. |
| [25] | Duan Y H, Wang Z Y, Fang Y, et al. 2024. A secreted fungal laccase targets the receptor kinase OsSRF3 to inhibit OsBAK1-OsSRF3- mediated immunity in rice. Nat Commun, 15(1): 7891. |
| [26] | Fabrice T N, Vogler H, Draeger C, et al. 2018. LRX proteins play a crucial role in pollen grain and pollen tube cell wall development. Plant Physiol, 176(3): 1981-1992. |
| [27] | Feng X Y, Li Q, Liu Y, et al. 2024. Evolutionary and immune- activating character analyses of NLR genes in algae suggest the ancient origin of plant intracellular immune receptors. Plant J, 119(5): 2316-2330. |
| [28] | Ferrari S, Galletti R, Vairo D, et al. 2006. Antisense expression of the Arabidopsis thaliana AtPGIP1 gene reduces polygalacturonase- inhibiting protein accumulation and enhances susceptibility to Botrytis cinerea. Mol Plant Microbe Interact, 19(8): 931-936. |
| [29] | Geng B H, Wang Q, Huang R S, et al. 2021. A novel LRR-RLK (CTLK) confers cold tolerance through regulation on the C- repeat-binding factor pathway, antioxidants, and proline accumulation. Plant J, 108(6): 1679-1689. |
| [30] | Gómez-Gómez L, Boller T. 2000. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell, 5(6): 1003-1011. |
| [31] | Harkenrider M, Sharma R, de Vleesschauwer D, et al. 2016. Overexpression of rice wall-associated kinase 25 (OsWAK25) alters resistance to bacterial and fungal pathogens. PLoS One, 11(1): e0147310. |
| [32] | He C M, Wang J, Dong R, et al. 2020. Overexpression of an antisense RNA of maize receptor-like kinase gene ZmRLK7 enlarges the organ and seed size of transgenic Arabidopsis plants. Front Plant Sci, 11: 579120. |
| [33] | He K, Gou X P, Yuan T, et al. 2007. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol, 17(13): 1109-1115. |
| [34] | Huang J, Wu X, Gao Z. 2021. The RING-type protein BOI negatively regulates the protein level of a CC-NBS-LRR in Arabidopsis. Biochem Biophys Res Commun, 578: 104-109. |
| [35] | Huang S J, Jia A L, Ma S C, et al. 2023. NLR signaling in plants: From resistosomes to second messengers. Trends Biochem Sci, 48(9): 776-787. |
| [36] | Hwang S G, Kim D S, Jang C S. 2011. Comparative analysis of evolutionary dynamics of genes encoding leucine-rich repeat receptor-like kinase between rice and Arabidopsis. Genetica, 139(8): 1023-1032. |
| [37] | Jehle A K, Lipschis M, Albert M, et al. 2013. The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell, 25(6): 2330-2340. |
| [38] | Jia M, Shen X Q, Tang Y, et al. 2021. A karyopherin constrains nuclear activity of the NLR protein SNC1 and is essential to prevent autoimmunity in Arabidopsis. Mol Plant, 14(10): 1733-1744. |
| [39] | Jubic L M, Saile S, Furzer O J, et al. 2019. Help wanted: Helper NLRs and plant immune responses. Curr Opin Plant Biol, 50: 82-94. |
| [40] | Kajava A V, Anisimova M, Peeters N. 2008. Origin and evolution of GALA-LRR, a new member of the CC-LRR subfamily: From plants to bacteria? PLoS One, 3(2): e1694. |
| [41] | Kalunke R M, Tundo S, Benedetti M, et al. 2015. An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front Plant Sci, 6: 146. |
| [42] | Kapos P, Devendrakumar K T, Li X. 2019. Plant NLRs: From discovery to application. Plant Sci, 279: 3-18. |
| [43] | Kileeg Z, Haldar A, Khan H, et al. 2023. Differential expansion and retention patterns of LRR-RLK genes across plant evolution. Plant Direct, 7(12): e556. |
| [44] | Kobe B, Kajava A V. 2001. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol, 11(6): 725-732. |
| [45] | Kourelis J, Marchal C, Kamoun S. 2023. NLR immune receptor- nanobody fusions confer plant disease resistance. Science, 379: 934-939. |
| [46] | Le M H, Cao Y, Zhang X C, et al. 2014. LIK1, a CERK1- interacting kinase, regulates plant immune responses in Arabidopsis. PLoS One, 9(7): e102245. |
| [47] | Li C Y, Wang L, Cui Y C, et al. 2016. Two FERONIA-like receptor (FLR) genes are required to maintain architecture, fertility, and seed yield in rice. Mol Breed, 36(11): 151. |
| [48] | Li J, Wen J Q, Lease K A, et al. 2002. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell, 110(2): 213-222. |
| [49] | Li T G, Wang B L, Yin C M, et al. 2019. The Gossypium hirsutum TIR-NBS-LRR gene GhDSC1 mediates resistance against Verticillium wilt. Mol Plant Pathol, 20(6): 857-876. |
| [50] | Li X L, Zheng Z Q, Kong X X, et al. 2019. A typical receptor kinase RINRK 1 required for rhizobial infection but not nodule development in Lotus japonicus. Plant Physiol, 181(2): 804-816. |
| [51] | Liu P L, Du L, Huang Y, et al. 2017. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol Biol, 17(1): 47. |
| [52] | Liu X S, Liang CC, Hou S G, et al. 2020. The LRR-RLK protein HSL3 regulates stomatal closure and the drought stress response by modulating hydrogen peroxide homeostasis. Front Plant Sci, 11: 548034. |
| [53] | Liu Y, Zeng Z, Zhang Y M, et al. 2021. An angiosperm NLR Atlas reveals that NLR gene reduction is associated with ecological specialization and signal transduction component deletion. Mol Plant, 14(12): 2015-2031. |
| [54] | Liu Y, Zhang Y M, Tang Y, et al. 2023. The evolution of plant NLR immune receptors and downstream signal components. Curr Opin Plant Biol, 73: 102363. |
| [55] | Luo Y C, Sangha J S, Wang S H, et al. 2012. Marker-assisted breeding of Xa4, Xa21 and Xa27 in the restorer lines of hybrid rice for broad-spectrum and enhanced disease resistance to bacterial blight. Mol Breed, 30(4): 1601-1610. |
| [56] | Lv L L, Liu Y S, Bai S H, et al. 2022. A TIR-NBS-LRR gene MdTNL1 regulates resistance to Glomerella leaf spot in apple. Int J Mol Sci, 23(11): 6323. |
| [57] | Ma X Y, Xu G Y, He P, et al. 2016. SERKing coreceptors for receptors. Trends Plant Sci, 21(12): 1017-1033. |
| [58] | Man J, Gallagher J P, Bartlett M. 2020. Structural evolution drives diversification of the large LRR-RLK gene family. New Phytol, 226(5): 1492-1505. |
| [59] | Martin R, Qi T, Zhang H, et al. 2020. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science, 370: eabd9993. |
| [60] | Masle J, Gilmore S R, Farquhar G D. 2005. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature, 436(7052): 866-870. |
| [61] | Matsushima N, Miyashita H. 2012. Leucine-rich repeat (LRR) domains containing intervening motifs in plants. Biomolecules, 2(2): 288-311. |
| [62] | Maulik A, Ghosh H, Basu S. 2009. Comparative study of protein- protein interaction observed in PolyGalacturonase-Inhibiting Proteins from Phaseolus vulgaris and Glycine max and PolyGalacturonase from Fusarium moniliforme. BMC Genomics, 10: S19. |
| [63] | Maulik A, Sarkar A I, Devi S, et al. 2012. Polygalacturonase-inhibiting proteins: Leucine-rich repeat proteins in plant defence. Plant Biol, 14: 22-30. |
| [64] | Nazar R N, Castroverde C D M, Xu X, et al. 2019. Wounding induces tomato Ve1 R-gene expression. Planta, 249(6): 1779-1797. |
| [65] | Ngou B P M, Wyler M, Schmid M W, et al. 2024. Evolutionary trajectory of pattern recognition receptors in plants. Nat Commun, 15: 308. |
| [66] | Ouyang S Q, Liu Y F, Liu P, et al. 2010. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J, 62(2): 316-329. |
| [67] | Pacheco J M, Song L M, Kuběnová L, et al. 2023. Cell surface receptor kinase FERONIA linked to nutrient sensor TORC signaling controls root hair growth at low temperature linked to low nitrate in Arabidopsis thaliana. New Phytol, 238(1): 169-185. |
| [68] | Park C H, Bi Y, Youn J H, et al. 2022. Deconvoluting signals downstream of growth and immune receptor kinases by phosphocodes of the BSU1 family phosphatases. Nat Plants, 8(6): 646-655. |
| [69] | Park S, Moon J C, Park Y C, et al. 2014. Molecular dissection of the response of a rice leucine-rich repeat receptor-like kinase (LRR-RLK) gene to abiotic stresses. J Plant Physiol, 171(17): 1645-1653. |
| [70] | Ravindran P, Yong S Y, Mohanty B, et al. 2020. An LRR-only protein regulates abscisic acid-mediated abiotic stress responses during Arabidopsis seed germination. Plant Cell Rep, 39(7): 909-920. |
| [71] | Ron M, Avni A. 2004. The receptor for the fungal elicitor ethylene- inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell, 16(6): 1604-1615. |
| [72] | Rose L E, Bittner-Eddy P D, Langley C H, et al. 2004. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics, 166( 3): 1517-1527. |
| [73] | Saijo Y, Loo E P I, Yasuda S. 2018. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J, 93(4): 592-613. |
| [74] | Salcedo A, Rutter W, Wang S C, et al. 2017. Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science, 358: 1604-1606. |
| [75] | Santiago J, Henzler C, Hothorn M. 2013. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science, 341: 889-892. |
| [76] | Sarazin V, Duclercq J, Mendou B, et al. 2015. Arabidopsis BNT1, an atypical TIR-NBS-LRR gene, acting as a regulator of the hormonal response to stress. Plant Sci, 239: 216-229. |
| [77] | Sarnowska E, Kubala S, Cwiek P, et al. 2023. A non-canonical function of Arabidopsis ERECTA proteins and a role of the SWI3B subunit of the SWI/SNF chromatin remodeling complex in gibberellin signaling. Plant J, 115(3): 788-802. |
| [78] | Saur I M L, Kadota Y, Sklenar J, et al. 2016. NbCSPR underlies age-dependent immune responses to bacterial cold shock protein in Nicotiana benthamiana. Proc Natl Acad Sci USA, 113(12): 3389-3394. |
| [79] | Sede A R, Borassi C, Wengier D L, et al. 2018. Arabidopsis pollen extensins LRX are required for cell wall integrity during pollen tube growth. FEBS Lett, 592(2): 233-243. |
| [80] | Shen Y, Diener A C. 2013. Arabidopsis thaliana resistance to fusarium oxysporum 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genet, 9(5): e1003525. |
| [81] | Sierla M, Hõrak H, Overmyer K, et al. 2018. The receptor-like pseudokinase GHR1 is required for stomatal closure. Plant Cell, 30(11): 2813-2837. |
| [82] | Song W, Liu L, Yu D L, et al. 2024. Substrate-induced condensation activates plant TIR domain proteins. Nature, 627: 847-853. |
| [83] | Song Y J, Niu R F, Yu H L, et al. 2022. OsSLA1 functions in leaf angle regulation by enhancing the interaction between OsBRI1 and OsBAK1 in rice. Plant J, 110(4): 1111-1127. |
| [84] | Tan X, Calderon-Villalobos L I A, Sharon M, et al. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature, 446: 640-645. |
| [85] | Torii K U. 2004. Leucine-rich repeat receptor kinases in plants: Structure, function, and signal transduction pathways. Int Rev Cytol, 234: 1-46. |
| [86] | Tornero P, Chao R A, Luthin W N, et al. 2002. Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell, 14(2): 435-450. |
| [87] | van der Burgh A M, Postma J, Robatzek S, et al. 2019. Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol Plant Pathol, 20(3): 410-422. |
| [88] | Walker J C. 1994. Structure and function of the receptor-like protein kinases of higher plants. Plant Mol Biol, 26(5): 1599-1609. |
| [89] | Wang J H, Kucukoglu M, Zhang L, et al. 2013. The Arabidopsis LRR-RLK, PXC1, is a regulator of secondary wall formation correlated with the TDIF-PXY/TDR-WOX4 signaling pathway. BMC Plant Biol, 13: 94. |
| [90] | Wang S, Wang X Q, Zhang R Y, et al. 2013. RppM, encoding a typical CC-NBS-LRR protein, confers resistance to southern corn rust in maize. Front Plant Sci, 13: 951318. |
| [91] | Wang X M, Cheng R, Xu D C, et al. 2023. MG1 interacts with a protease inhibitor and confers resistance to rice root-knot nematode. Nat Commun, 14(1): 3354. |
| [92] | Wang Z C, Yang D W, Zhong G T, et al. 2024. Nucleotide-binding leucine-rich repeat receptor homologs Pib and PibH8 interact and contribute to immunity in rice. Plant Physiol, 195(4): 3010-3023. |
| [93] | Wu F H, Chi Y, Jiang Z H, et al. 2020. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature, 578: 577-581. |
| [94] | Wu T, Peng C E, Li B B, et al. 2019. OsPGIP1-mediated resistance to bacterial leaf streak in rice is beyond responsive to the polygalacturonase of Xanthomonas oryzae pv. oryzicola. Rice, 12(1): 90. |
| [95] | Xia C X, Liang G H, Chong K, et al. 2023. The COG1-OsSERL2 complex senses cold to trigger signaling network for chilling tolerance in japonica rice. Nat Commun, 14(1): 3104. |
| [96] | Xiao G, Laksanavilat N, Cesari S, et al. 2024. The unconventional resistance protein PTR recognizes the Magnaporthe oryzae effector AVR-Pita in an allele-specific manner. Nat Plants, 10(6): 994-1004. |
| [97] | Xie Y J, Wang Y P, Yu X Z, et al. 2022. SH3P2, an SH3 domain- containing protein that interacts with both Pib and AvrPib, suppresses effector-triggered, Pib-mediated immunity in rice. Mol Plant, 15(12): 1931-1946. |
| [98] | Xing L, Hu P, Liu J, et al. 2018. Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol Plant, 11(6): 874-878. |
| [99] | Yang C, Liu R, Pang J, et al. 2021. Poaceae-specific cell wall- derived oligosaccharides activate plant immunity via OsCERK1 during Magnaporthe oryzae infection in rice. Nat Commun, 12(1): 2178. |
| [100] | Yang J, Ji L X, Zhu B H, et al. 2018. OsCML16 interacts with a novel CC-NBS-LRR protein OsPi304 in the Ca2+/Mg2+ dependent and independent manner in rice. Biochem Biophys Res Commun, 504(1): 346-351. |
| [101] | Yin J L, Wang L Q, Jin T T, et al. 2021. A cell wall-localized NLR confers resistance to Soybean mosaic virus by recognizing viral- encoded cylindrical inclusion protein. Mol Plant, 14(11): 1881-1900. |
| [102] | Yin Z Y, Wang N, Pi L, et al. 2021. Nicotiana benthamiana LRR- RLP NbEIX2 mediates the perception of an EIX-like protein from Verticillium dahliae. J Integr Plant Biol, 63(5): 949-960. |
| [103] | Zhang H H, Chen C H, Li L L, et al. 2021. A rice LRR receptor- like protein associates with its adaptor kinase OsSOBIR1 to mediate plant immunity against viral infection. Plant Biotechnol J, 19(11): 2319-2332. |
| [104] | Zhang Q, Yan Q, Yuan X, et al. 2021. Two polygalacturonase- inhibiting proteins (VrPGIP) of Vigna radiata confer resistance to bruchids (Callosobruchus spp.). J Plant Physiol, 258/259: 153376. |
| [105] | Zhao B, Ardales E Y, Raymundo A, et al. 2004. The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv. oryzicola confers a nonhost defense reaction on maize with resistance gene Rxo1. Mol Plant Microbe Interact, 17(7): 771-779. |
| [106] | Zhao C Z, Zayed O, Yu Z P, et al. 2018. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc Natl Acad Sci USA, 115(51): 13123-13128. |
| [107] | Zhao M C, Tang S, Zhang H S, et al. 2020. DROOPY LEAF1 controls leaf architecture by orchestrating early brassinosteroid signaling. Proc Natl Acad Sci USA, 117(35): 21766-21774. |
| [108] | Zhao M C, Zhang Q, Liu H, et al. 2023. The osmotic stress- activated receptor-like kinase DPY1 mediates SnRK2 kinase activation and drought tolerance in Setaria. Plant Cell, 35(10): 3782-3808. |
| [109] | Zhao Q Q, Bao J L, Li H L, et al. 2024. Structural and biochemical basis of FLS2-mediated signal activation and transduction in rice. Plant Commun, 5(3): 100785. |
| [110] | Zhou T, Wang Y, Chen J Q, et al. 2004. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Genet Genomics, 271(4): 402-415. |
| [111] | Zorzatto C, Machado J P, Lopes K V, et al. 2015. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature, 520: 679-682. |
| [1] | Xia Xiaodong, Zhang Xiaobo, Wang Zhonghao, Cheng Benyi, Sun Huifeng, Xu Xia, Gong Junyi, Yang Shihua, Wu Jianli, Shi Yongfeng, Xu Rugen. Mapping and Functional Analysis of LE Gene in a Lethal Etiolated Rice Mutant at Seedling Stage [J]. Rice Science, 2023, 30(6): 567-576. |
| [2] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [3] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| [4] | Jiang Changjie, Liang Zhengwei, Xie Xianzhi. Priming for Saline-Alkaline Tolerance in Rice: Current Knowledge and Future Challenges [J]. Rice Science, 2023, 30(5): 417-425. |
| [5] | Sheikh Faruk Ahmed, Hayat Ullah, May Zun Aung, Rujira Tisarum, Suriyan Cha-Um, Avishek Datta. Iron Toxicity Tolerance of Rice Genotypes in Relation to Growth, Yield and Physiochemical Characters [J]. Rice Science, 2023, 30(4): 321-334. |
| [6] | Van Quoc Giang, Huynh Ky, Nguyen Chau Thanh Tung, Nguyen Loc Hien, Nguyen van Manh, Nguyen Nhut Thanh, Vo Cong Thanh, Swee Keong Yeap. Novel Deletion in Exon 7 of Betaine Aldehyde Dehydrogenase 2 (BADH2) [J]. Rice Science, 2023, 30(2): 104-112. |
| [7] | Pandia Rajan Jeyaraj, Siva Prakash Asokan, Edward Rajan Samuel Nadar. Computer-Assisted Real-Time Rice Variety Learning Using Deep Learning Network [J]. Rice Science, 2022, 29(5): 489-498. |
| [8] | Wang Chenjiaozi, Zhao Mei, Shu Canwei, Zhou Erxun. Three Genes Related to Trehalose Metabolism Affect Sclerotial Development of Rhizoctonia solani AG-1 IA, Causal Agent of Rice Sheath Blight [J]. Rice Science, 2022, 29(3): 268-276. |
| [9] | Hyeran Moon, Young-Ah Kim, Ryoung Shin, Chang-Jin Park. Nucleus-Encoded Thylakoid Protein, OsY3IP1, Confers Enhanced Tolerance to Saline and Alkaline Stresses in Rice [J]. Rice Science, 2022, 29(3): 225-236. |
| [10] | Chen Wei, Cai Yicong, Shakeel Ahmad, Wang Yakun, An Ruihu, Tang Shengjia, Guo Naihui, Wei Xiangjin, Tang Shaoqing, Shao Gaoneng, Jiao Guiai, Xie Lihong, Hu Shikai, Sheng Zhonghua, Hu Peisong. NRL3 Interacts with OsK4 to Regulate Heading Date in Rice [J]. Rice Science, 2022, 29(3): 237-246. |
| [11] | Dan Zeng, Chunchao Wang, Junpin Xie, Fan Zhang, Jialing Lu, Xiaorong Shi, Yingyao Shi, Yongli Zhou. Stress-Activated Protein Kinase OsSAPK7 Regulates Salt- Stress Tolerance by Modulating Diverse Stress-Defensive Responses in Rice [J]. Rice Science, 2021, 28(6): 547-556. |
| [12] | Tao Wang, Lijuan Lou, Zeyu Li, Lianguang Shang, Quan Wang. Cloning and Characterization of Protein Prenyltransferase Alpha Subunit in Rice [J]. Rice Science, 2021, 28(6): 557-566. |
| [13] | Punia Sneh, Kumar Manoj, Kumar Siroha Anil, Singh Purewal Sukhvinder. Rice Bran Oil: Emerging Trends in Extraction, Health Benefit, and Its Industrial Application [J]. Rice Science, 2021, 28(3): 217-232. |
| [14] | Saiful Islam Md. Sensing and Uptake of Nitrogen in Rice Plant: A Molecular View [J]. Rice Science, 2019, 26(6): 343-355. |
| [15] | Lei He, Guang Chen, Sen Zhang, Zhennan Qiu, Jiang Hu, Dali Zeng, Guangheng Zhang, Guojun Dong, Zhenyu Gao, Deyong Ren, Lan Shen, Longbiao Guo, Qian Qian, Li Zhu. Functional Analysis of Three Rice Chloroplast Transit Peptides [J]. Rice Science, 2019, 26(1): 11-20. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||