
Rice Science ›› 2021, Vol. 28 ›› Issue (5): 442-456.DOI: 10.1016/j.rsci.2021.07.005
• Research Paper • Previous Articles Next Articles
Ersong Zheng1,2,#, Xuming Wang1,#, Rumeng Xu1,2, Feibo Yu3, Chao Zheng1,4, Yong Yang1, Yang Chen1, Jianping Chen1,5, Chengqi Yan6( ), Jie Zhou1(
), Jie Zhou1( )
)
Received:2020-06-15
Accepted:2020-10-10
Online:2021-09-28
Published:2021-09-28
About author:#These authors contributed equally to this work
Ersong Zheng, Xuming Wang, Rumeng Xu, Feibo Yu, Chao Zheng, Yong Yang, Yang Chen, Jianping Chen, Chengqi Yan, Jie Zhou. Regulation of OsPR10a Promoter Activity by Phytohormone and Pathogen Stimulation in Rice[J]. Rice Science, 2021, 28(5): 442-456.
Add to citation manager EndNote|Ris|BibTeX

Fig. S1. Map of the OsPR10a::GUS vector used for transformation. LB, Left border; RB, Right border; Pnos, Nos promoter; HPT, Hygromycin phosphotransferase gene; T35S:CaMV35S terminator; GUS, β-glucuronidase gene; Tnos, Nos terminator.
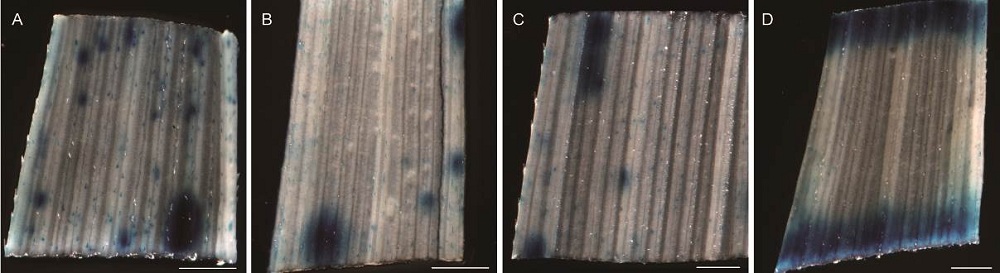
Fig. S2. Histochemical staining of OsPR10a::GUS expression in leves of T0 rice transgenic plants.A, B and C, GUS expression with weak or strong blue spots scattered randomly on the leaf surface of three independent transgenic lines. D, GUS expression only induced at the cutting edges. Scale bars, 0.5 mm.
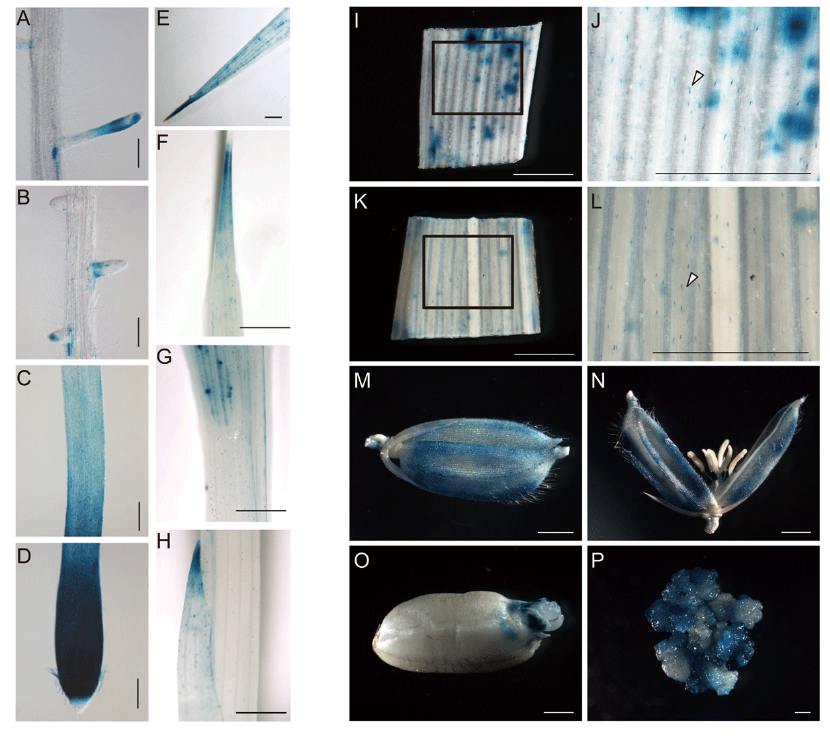
Fig. 1. Characterization of tissue expression pattern of OsPR10a.A?H, Histochemical staining of the OsPR10a::GUS activity in the 5-day-old rice transgenic plants. Parts of the root and shoot, including mature region with lateral roots (A and B), elongation zone (C), root meristem (D), the first true leaf (E), semi true leaf (F and G) and coleoptile (H) with GUS staining were shown. Scale bars are 200 μm in A to D and 1 mm in E to H, respectively.I?P, GUS expression in the young leaf of 14-day-old seedling (I and J), flag leaf (K and L), floret before flowering (M and N), germinating seed (O) and resistant callus (P). The areas indicated by rectangles in I and K were enlarged to show the GUS activity in the trichomes on the surface of leaf (J and L) as indicated by white arrows. Scale bars, 1 mm.
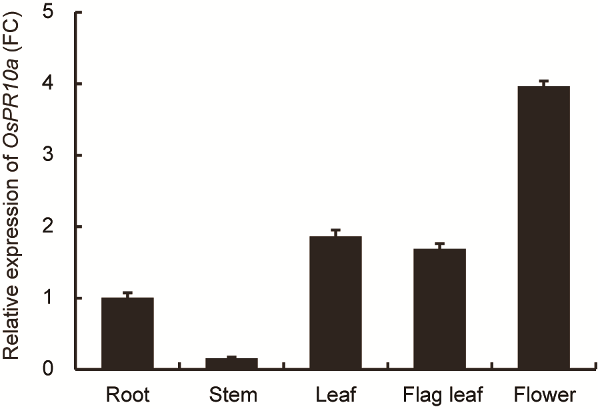
Fig. S3. OsPR10a gene expression in tissues and organs of wild type plants.OsPR10a gene expression was analyzed in different tissues and organs of wild type plant by qRT-PCR. The expression level was represented as fold change (FC) relative to the level of root was shown. Data are Mean?± SD (n?= 3).
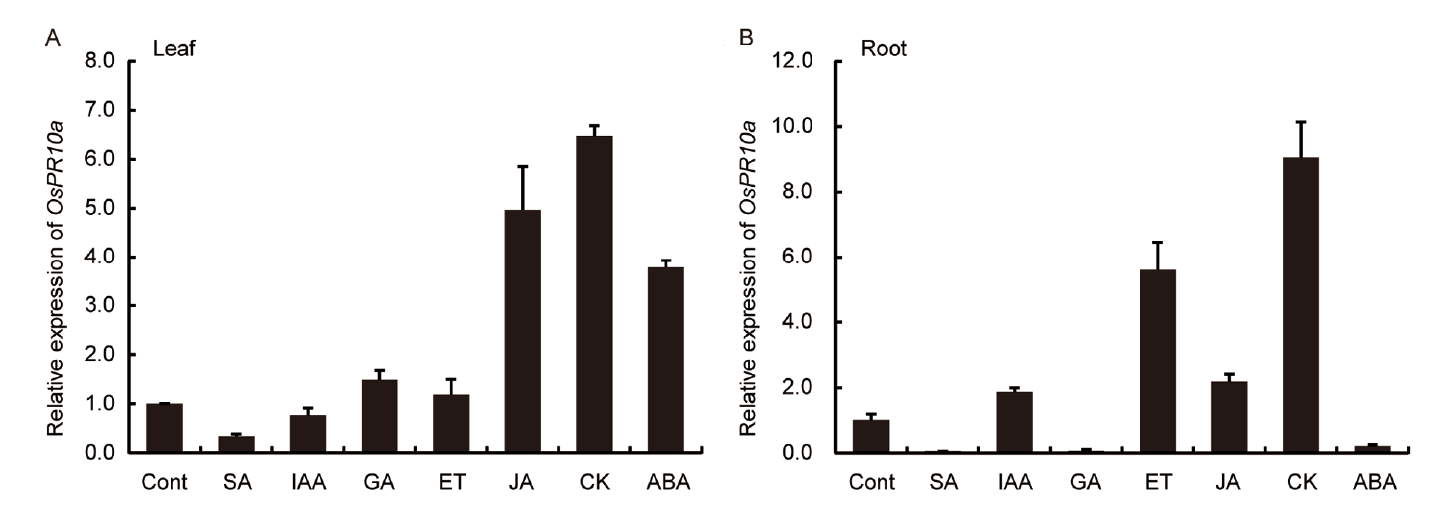
Fig. 2. Expression of OsPR10a mRNA transcript level in response to hormone treatments for 24 h compared to the control (Cont).A, Expression in leaf. B, Expression in root. Data are Mean ± SD (n = 3).SA, Salicylic acid; IAA, Indoleacetic acid; GA, Gibberellin; ET, Ethylene; JA, Jasmonic acid; CK, Cytokinin; ABA, Abscisic acid.
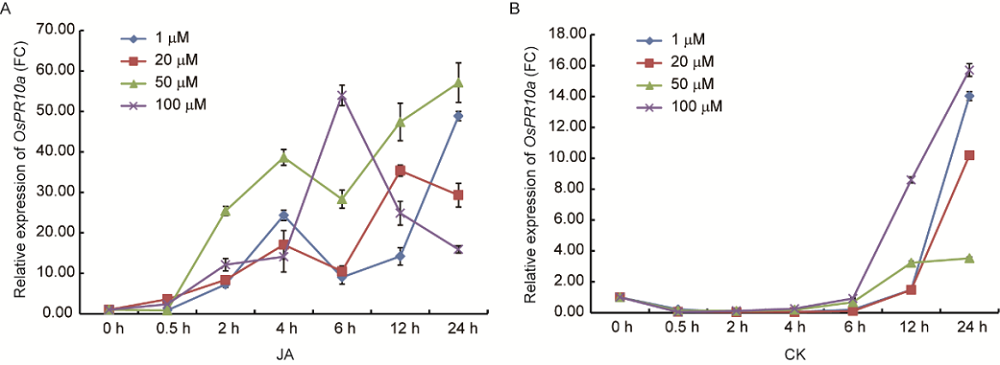
Fig. S4. Dynamic changes of OsPR10a gene transcript levels in response to JA and CK treatments under different concentrations and time periods.A, OsPR10a gene expression in response to MeJA treatment. B, OsPR10a gene expression in response to KN treatment. The OsPR10a expression level was represented as fold change (FC) relative to the value at 0 h.
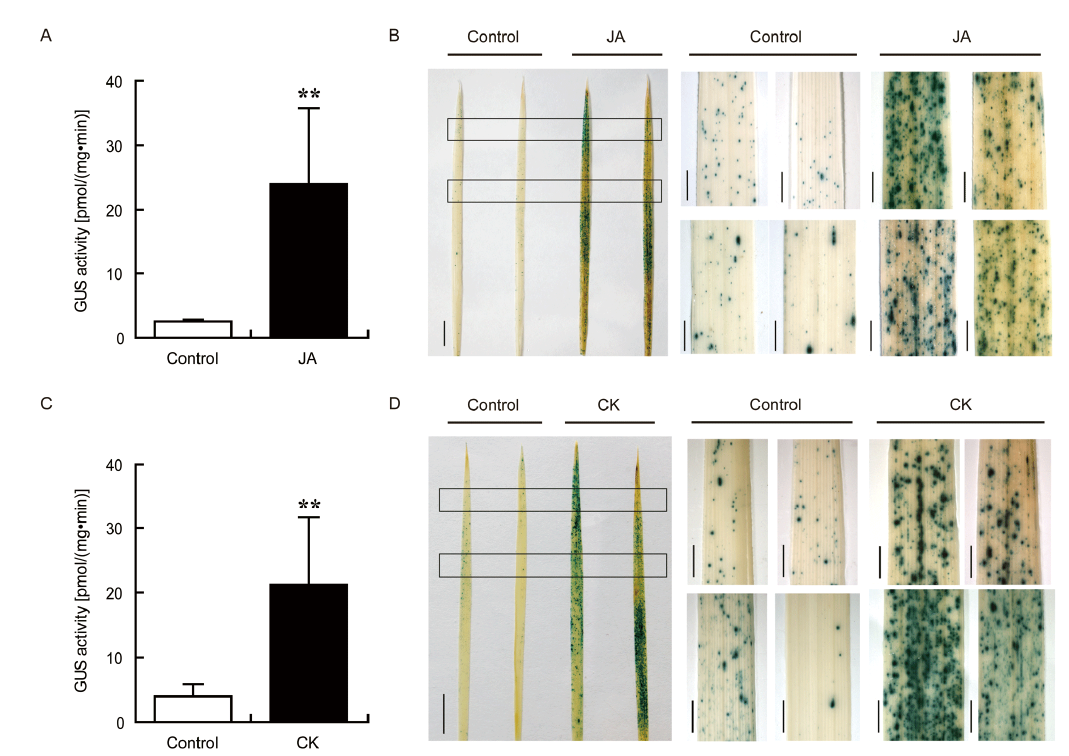
Fig. 3. Expression of OsPR10a::GUS in response to jasmonate (JA) and cytokinin (CK) treatments.A and C, GUS activity in the leaves of OsPR10a::GUS transgenic plants treated with JA (100 μmol/L) and CK (100 μmol/L) for 24 h. Data are shown as Mean ± SD (n = 3). ** indicates significant difference at the 0.01 level by the Student’s t-test. B and D, GUS staining in the leaves of OsPR10::GUS transgenic plants treated with JA (100 μmol/L) and CK (100 μmol/L) for 24 h. Two entire leaves for each treatment were shown. Scale bars, 1 cm. Magnified images of the areas with rectangles were shown aside. Scale bars, 1 mm.
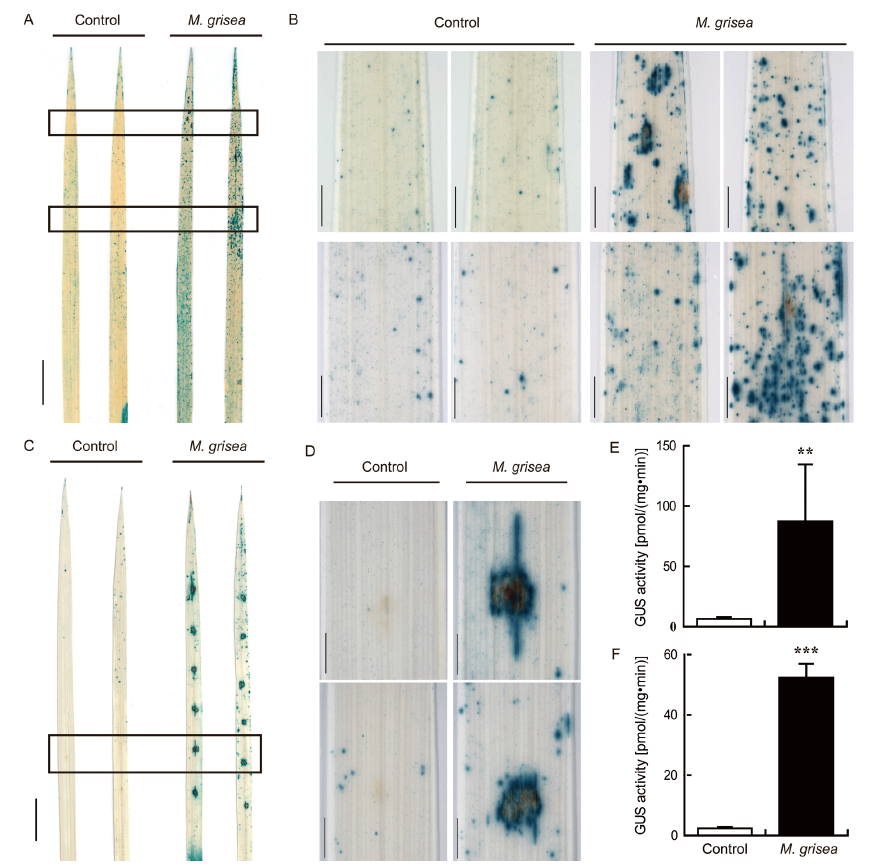
Fig. 4. Expression of OsPR10a::GUS in response to M. grisea infection.A, GUS staining in leaves of OsPR10a::GUS transgenic plants sprayed with H2O + 0.05% Tween 20 (Control) or M. grisea strain Guy 11 for 3 d. Two entire leaves for each treatment were shown. Scale bar, 1 cm.B, Magnified images of the area indicated by rectangles in A. Scale bars, 1 mm.C, GUS staining in detached leaves of OsPR10a::GUS transgenic plants spot inoculated with H2O + 0.05% Tween 20 (Control) or M. grisea strain Guy 11 for 2 d. Two entire leaves for each treatment were shown. Scale bar, 1 cm.D, Magnified images of the area indicated by rectangles in C. Scale bars, 1 mm.E and F, GUS activities in leaves of OsPR10a::GUS transgenic plants sprayed (E) or spot inoculated (H) with H2O + 0.05% Tween 20 (Control) or M. grisea strain Guy 11. Data are shown as Mean ± SD (n = 3). ** and *** indicate significant differences at the 0.01 and 0.001 levels by the Student’s t-test.
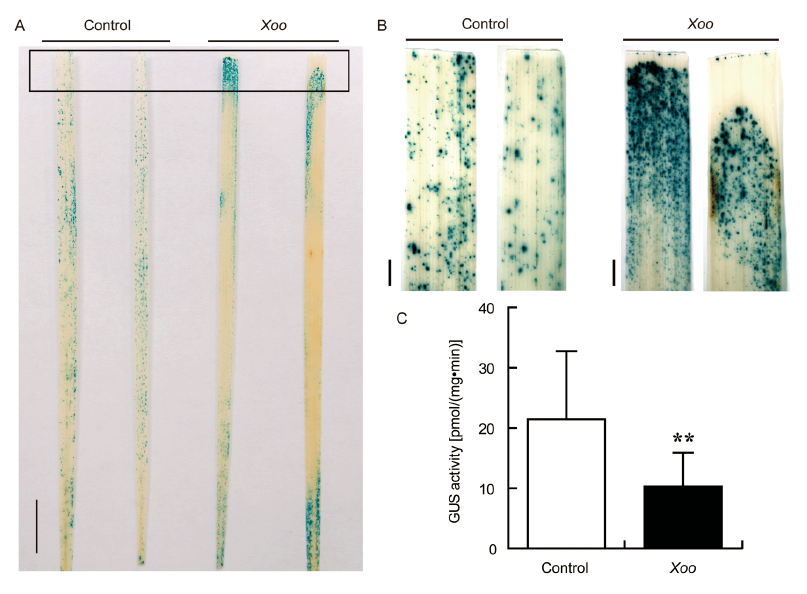
Fig. 5. Expression of OsPR10a::GUS in response to Xoo infection.A, GUS staining in leaves of OsPR10a::GUS transgenic plants with clip inoculation by H2O (Control) or Xoo strain PXO341 for 24 h. Two entire leaves for each treatment were shown. Scale bar, 1 cm. B, Magnified images of the inoculated area indicated by the rectangle in A. Scale bars, 1 mm. C, GUS activity in leaves of OsPR10a::GUS transgenic plants with clip inoculation by H2O (Control) or Xoo strain PXO341 for 24 h. Data are shown as Mean ± SD (n = 3). ** indicates a significant difference at the 0.01 level by the Student’s t-test.
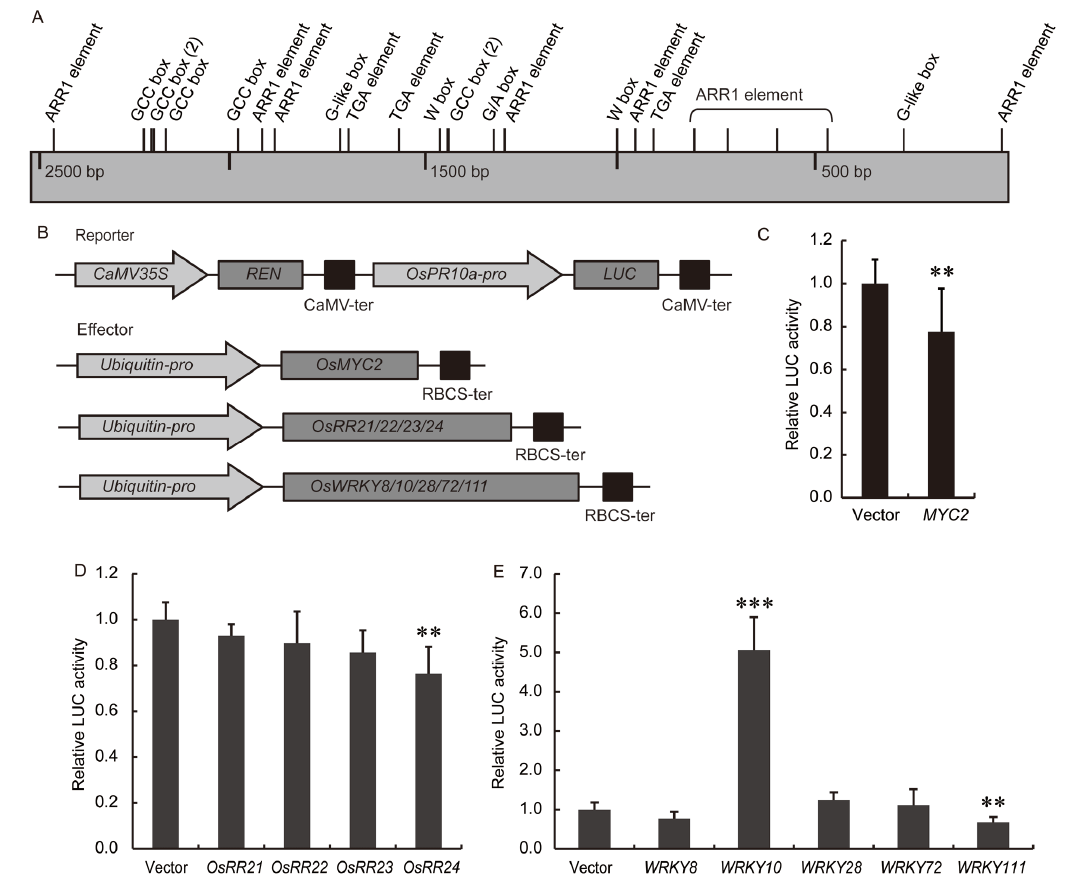
Fig. 6. Transactivation assays of OsPR10a promoter on JA and CK responsive transcription factors.A, Schematic diagram of OsPR10a promoter (2 523 bp). The numbers indicate the distance from the start codon. The predicted cis-elements (GCC box, G-like box, G/A box, ARR1 element, W box and TGA element) are indicated on the forward DNA strand. B, Schematic diagram of double-reporter and effector plasmids used in the dual luciferase (LUC) assay. The OsPR10a promoter was fused to firefly LUC and the renilla luciferase (REN) driven by CaMV35S was served as an internal control. The effector plasmids contain the OsMYC2, OsRR21/22/ 23/24 and OsWRKY8/10/28/72/111 genes driven by Ubiquitin, respectively. C?E, Transient transcriptional activities of OsMYC2 (C), OsRR21/22/23/24 (D) and OsWRKY8/10/28/72/111 (E) on the expression of OsPR10a promoter. The relative LUC activity was shown as fold change to the empty vector whose LUC/REN ratio was set to 1. All data are shown as Mean ± SD (n = 6). ** and *** indicate significant differences at the 0.01 and 0.001 levels by the Student’s t-test.
| Probe Set ID | RAP_Locus | Gene symbol | Fold Change (FC) |
|---|---|---|---|
| Os.29979.1.S1_at | Os01g0186000 | OsWRKY10 | 9.7 |
| Os.30512.1.S1_at | Os01g0626400 | OsWRKY11 | 2.8 |
| Os.30386.1.S1_at | Os01g0734000 | OsWRKY23 | 2.7 |
| Os.11773.1.S1_at | Os05g0343400 | OsWRKY53 | 2.0 |
| Os.14882.1.S1_at | Os12g0116600 | OsWRKY56 | 4.0 |
| Os.50015.1.S1_at | Os06g0649000 | OsWRKY28 | 9.6 |
| Os.27227.1.S1_at | Os09g0334500 | OsWRKY74 | 2.3 |
| Os.49656.1.S1_at | Os04g0287400 | OsWRKY51 | 2.2 |
| Os.33990.1.S1_at | Os11g0116900 | OsWRKY46 | 3.6 |
| Os.33131.1.A1_at | Os03g0321700 | OsWRKY55 | 3.9 |
| Os.55827.1.S1_at | Os11g0490900 | OsWRKY72 | 13.2 |
| Os.29987.2.S1_at | Os05g0583000 | OsWRKY8 | 21.9 |
| Os.2160.2.S1_x_at | Os01g0750100 | OsWRKY13 | 2.2 |
| Os.12032.1.S1_at | Os02g0181300 | OsWRKY71 | 6.9 |
| OsAffx.11050.2.S1_x_at | Os01g0246700 | OsWRKY1 | 6.4 |
Table S1 List of OsWRKY genes induced greater than 2-fold in leaves of wild type rice plants treated with 100 μmol/L MeJA.
| Probe Set ID | RAP_Locus | Gene symbol | Fold Change (FC) |
|---|---|---|---|
| Os.29979.1.S1_at | Os01g0186000 | OsWRKY10 | 9.7 |
| Os.30512.1.S1_at | Os01g0626400 | OsWRKY11 | 2.8 |
| Os.30386.1.S1_at | Os01g0734000 | OsWRKY23 | 2.7 |
| Os.11773.1.S1_at | Os05g0343400 | OsWRKY53 | 2.0 |
| Os.14882.1.S1_at | Os12g0116600 | OsWRKY56 | 4.0 |
| Os.50015.1.S1_at | Os06g0649000 | OsWRKY28 | 9.6 |
| Os.27227.1.S1_at | Os09g0334500 | OsWRKY74 | 2.3 |
| Os.49656.1.S1_at | Os04g0287400 | OsWRKY51 | 2.2 |
| Os.33990.1.S1_at | Os11g0116900 | OsWRKY46 | 3.6 |
| Os.33131.1.A1_at | Os03g0321700 | OsWRKY55 | 3.9 |
| Os.55827.1.S1_at | Os11g0490900 | OsWRKY72 | 13.2 |
| Os.29987.2.S1_at | Os05g0583000 | OsWRKY8 | 21.9 |
| Os.2160.2.S1_x_at | Os01g0750100 | OsWRKY13 | 2.2 |
| Os.12032.1.S1_at | Os02g0181300 | OsWRKY71 | 6.9 |
| OsAffx.11050.2.S1_x_at | Os01g0246700 | OsWRKY1 | 6.4 |
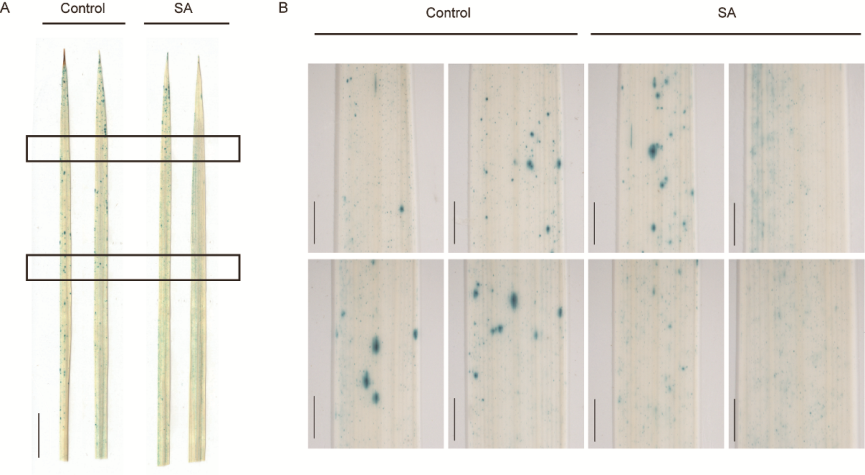
Fig. S6. Expression of OsPR10a::GUS in response to SA treatment. A, GUS staining in leaves of OsPR10::GUS transgenic plants with no treatment (control) or treated with SA (100 μmol/L) for 24 h. Two entire leaves for each treatment were shown. Scale bars, 1 cm. B, Magnified images of the areas with rectanges were shown aside. Scale bars, 1 mm.
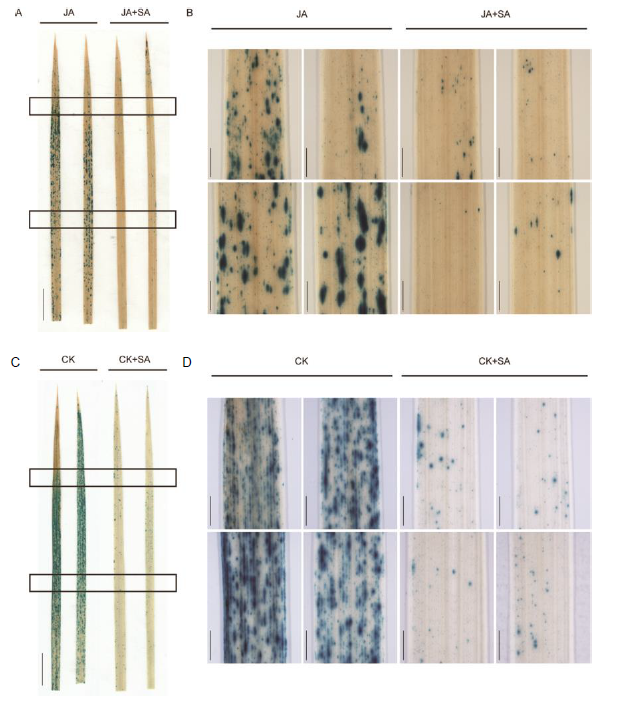
Fig. S7. Expression of OsPR10a::GUS in response to JA+SA treatment. A, GUS staining in leaves of OsPR10::GUS transgenic plants with MeJA (100 μmol/L) treatment alone or co-treatment with SA (100 μmol/L) for 24 h. Two entire leaves for each treatment were shown. Scale bars, 1 cm. B, Magnified images of the areas with rectanges were shown aside. Scale bars, 1 mm. C, GUS staining in leaves of OsPR10::GUS transgenic plants with KN (100 μmol/L) treatment alone or co-treatment with SA (100 μmol/L) for 24 h. Two entire leaves for each treatment were shown. Scale bars, 1 cm. D, Magnified images of the areas with rectanges were shown aside. Scale bars, 1 mm.
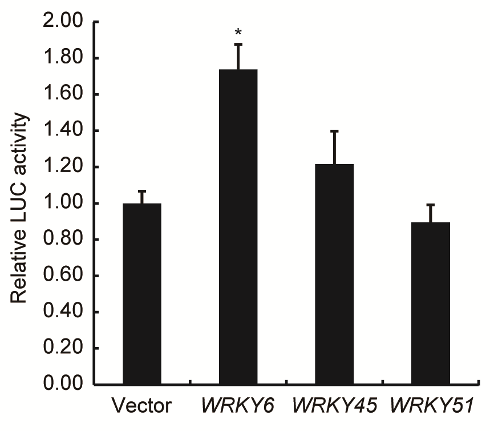
Fig. S8. The transcriptional effects of several Xoo responsive OsWRKY genes on the regulation of OsPR10a promoter.The transient transcriptional activity of OsWRKY6/45/51 on the expression of OsPR10a promoter. The relative LUC activity of each effector was shown as fold change to the empty vector whose LUC/REN ratio was set to 1. All data are shown as Mean ± SD (n = 6). * indicate the significant difference at P<0.01 by Student’s t-test analysis.
| Primer | Sequence (5'-3') |
|---|---|
| OsPR10a-Pr-F1 | AGGATCCATGCGTTTTTGCTGTAGGA |
| OsPR10a-Pr-R1 | AGGTACCCACTGAAGATATAATCTAACTAG |
| OsPR10a-Pr-F2 | GCAGCCCGGGGGATCCATGCGTTTTTGCTGTAGGAC |
| OsPR10a-Pr-R2 | ACCGCGGTGGCGGCCGCCACTGAAGATATAATCTAACTAGCT |
| OsMYC2-CDS-F1 | ATGAACCTTTGGACGGACGACAACG |
| OsMYC2-CDS-R1 | TTACCGGGCGGCGGTGCCA |
| OsMYC2-CDS-F2 | TATCAAGCTAATTCGAGCTCATGAACCTTTGGACGGACGACAACG |
| OsMYC2-CDS-R2 | CGACTCTAGAGGATCCTTACCGGGCGGCGGTGCC |
| OsRR21-CDS-F1 | ATGGCGCCGGTGGAGGATG |
| OsRR21-CDS-R1 | TCACATCTGTCCACTAAATCCGAAAATAT |
| OsRR21-CDS-F2 | GCTAATTCGAGCTCGGTACCATGGCGCCGGTGGAGGAT |
| OsRR21-CDS-R2 | CGACTCTAGAGGATCCTCACATCTGTCCACTAAATC |
| OsRR22-CDS-F1 | ATGCTTCTGGGTGCTTTGAGGATG |
| OsRR22-CDS-R1 | TCATATGCAGGCACCAAGTGGAAA |
| OsRR22-CDS-F2 | GCTAATTCGAGCTCGGTACCATGCTTCTGGGTGCTTTGA |
| OsRR22-CDS-R2 | CGACTCTAGAGGATCCTCATATGCAGGCACCAAGT |
| OsRR23-CDS-F1 | ATGAGGGCGGCGGAGGAGAGGAAG |
| OsRR23-CDS-R1 | TCATATGCAAGCTCCAAGGGAGTAGAAA |
| OsRR23-CDS-F2 | GCTAATTCGAGCTCGATGAGGGCGGCGGAGGAG |
| OsRR23-CDS-R2 | CGACTCTAGAGGATCTCATATGCAAGCTCCAAGG |
| OsRR24-CDS-F1 | ATGACGGTGGAGGAGAGGCAGGG |
| OsRR24-CDS-R1 | CTAGACCAGCTCCCAGTCCCCATC |
| OsRR24-CDS-F2 | GCTAATTCGAGCTCGGTACCATGACGGTGGAGGAGAGG |
| OsRR24-CDS-R2 | CGACTCTAGAGGATCCCTAGACCAGCTCCCAGTC |
| OsWRKY8-CDS2-F1 | ATGTCTGGACCTGGAGGAGGA |
| OsWRKY8-CDS2-R1 | TCACGGAAGGAAGAGGTCTTG |
| OsWRKY8-CDS2-F2 | TATCAAGCTAATTCGAGCTCATGTCTGGACCTGGAGGAGGAG |
| OsWRKY8-CDS2-R2 | CTAGAGGATCCCCGGGTACCTCACGGAAGGAAGAGGTCTTGC |
| OsWRKY10-CDS-F1 | ATGGCGGCTTCGCTGGGAC |
| OsWRKY10-CDS-R1 | TCAGAACGACGATTCCGACGAGT |
| OsWRKY10-CDS-F2 | TATCAAGCTAATTCGAGCTCATGGCGGCTTCGCTGGGAC |
| OsWRKY10-CDS-R2 | CTAGAGGATCCCCGGGTACCTCAGAACGACGATTCCGACGAG |
| OsWRKY28-CDS-F1 | ATGACCGCCGCGCCGGGG |
| OsWRKY28-CDS-R1 | TCAGTTCTTGGTCGGCGA |
| OsWRKY28-CDS-F2 | GCTAATTCGAGCTCGGTACCATGACCGCCGCGCCGGGG |
| OsWRKY28-CDS-R2 | CGACTCTAGAGGATCCTCAGTTCTTGGTCGGCGA |
| OsWRKY72-CDS-F1 | ATGGAGAACTTCCCGATACTCTT |
| OsWRKY72-CDS-R1 | CTACTGGAACATGTGGGAAGC |
| OsWRKY72-CDS-F2 | TATCAAGCTAATTCGAGCTCATGGAGAACTTCCCGATACTC |
| OsWRKY72-CDS-R2 | CTAGAGGATCCCCGGGTACCCTACTGGAACATGTGGGAAGC |
| OsWRKY111-CDS-F1 | ATGACCATCACACACGCTAG |
| OsWRKY111-CDS-R1 | CTAGAGCGACAGCCGGCGTCCA |
| OsWRKY111-CDS-F2 | TATCAAGCTAATTCGAGCTCATGACCATCACACACGCTAGC |
| OsWRKY111-CDS-R2 | CTAGAGGATCCCCGGGTACCCTAGAGCGACAGCCGGCG |
| OsWRKY6-CDS-F1 | ATGGATGGGGTGGAGGAG |
| OsWRKY6-CDS-R1 | TCAGGTCTGGGGATTAGCGG |
| OsWRKY6-CDS-F2 | TATCAAGCTAATTCGAGCTCATGGATGGGGTGGAGGAG |
| OsWRKY6-CDS-R2 | CTAGAGGATCCCCGGGTACCTCAGGTCTGGGGATTAGCGG |
| OsWRKY45-CDS-F1 | ATGACGTCATCGATGTCGC |
| OsWRKY45-CDS-R1 | TCAAAAGCTCAAACCCATAATGTCGT |
| OsWRKY45-CDS-F2 | TATCAAGCTAATTCGAGCTCATGACGTCATCGATGTCGC |
| OsWRKY45-CDS-R2 | CTAGAGGATCCCCGGGTACCTCAAAAGCTCAAACCCATAATGT |
| OsWRKY51-CDS-F1 | ATGATTACCATGGATCTGATGGGTGGG |
| OsWRKY51-CDS-R1 | TCAGGCGAGCGGCAGCGCCAG |
| OsWRKY51-CDS-F2 | TATCAAGCTAATTCGAGCTCATGATTACCATGGATCTGATGGGTGGG |
| OsWRKY51-CDS-R2 | CTAGAGGATCCCCGGGTACCTCAGGCGAGCGGCAGCGCCAG |
| OsPR10a-RT-F | GGGCACCATCTACACCATGAA |
| OsPR10a-RT-R | TCGTACTCCACCTTGAGCTT |
| OsAct1-RT-F | GAGTATGATGAGTCGGGTCCAG |
| OsAct1-RT-R | ACACCAACAATCCCAAACAGAG |
Table S2 Sequences of the primers used in this study.
| Primer | Sequence (5'-3') |
|---|---|
| OsPR10a-Pr-F1 | AGGATCCATGCGTTTTTGCTGTAGGA |
| OsPR10a-Pr-R1 | AGGTACCCACTGAAGATATAATCTAACTAG |
| OsPR10a-Pr-F2 | GCAGCCCGGGGGATCCATGCGTTTTTGCTGTAGGAC |
| OsPR10a-Pr-R2 | ACCGCGGTGGCGGCCGCCACTGAAGATATAATCTAACTAGCT |
| OsMYC2-CDS-F1 | ATGAACCTTTGGACGGACGACAACG |
| OsMYC2-CDS-R1 | TTACCGGGCGGCGGTGCCA |
| OsMYC2-CDS-F2 | TATCAAGCTAATTCGAGCTCATGAACCTTTGGACGGACGACAACG |
| OsMYC2-CDS-R2 | CGACTCTAGAGGATCCTTACCGGGCGGCGGTGCC |
| OsRR21-CDS-F1 | ATGGCGCCGGTGGAGGATG |
| OsRR21-CDS-R1 | TCACATCTGTCCACTAAATCCGAAAATAT |
| OsRR21-CDS-F2 | GCTAATTCGAGCTCGGTACCATGGCGCCGGTGGAGGAT |
| OsRR21-CDS-R2 | CGACTCTAGAGGATCCTCACATCTGTCCACTAAATC |
| OsRR22-CDS-F1 | ATGCTTCTGGGTGCTTTGAGGATG |
| OsRR22-CDS-R1 | TCATATGCAGGCACCAAGTGGAAA |
| OsRR22-CDS-F2 | GCTAATTCGAGCTCGGTACCATGCTTCTGGGTGCTTTGA |
| OsRR22-CDS-R2 | CGACTCTAGAGGATCCTCATATGCAGGCACCAAGT |
| OsRR23-CDS-F1 | ATGAGGGCGGCGGAGGAGAGGAAG |
| OsRR23-CDS-R1 | TCATATGCAAGCTCCAAGGGAGTAGAAA |
| OsRR23-CDS-F2 | GCTAATTCGAGCTCGATGAGGGCGGCGGAGGAG |
| OsRR23-CDS-R2 | CGACTCTAGAGGATCTCATATGCAAGCTCCAAGG |
| OsRR24-CDS-F1 | ATGACGGTGGAGGAGAGGCAGGG |
| OsRR24-CDS-R1 | CTAGACCAGCTCCCAGTCCCCATC |
| OsRR24-CDS-F2 | GCTAATTCGAGCTCGGTACCATGACGGTGGAGGAGAGG |
| OsRR24-CDS-R2 | CGACTCTAGAGGATCCCTAGACCAGCTCCCAGTC |
| OsWRKY8-CDS2-F1 | ATGTCTGGACCTGGAGGAGGA |
| OsWRKY8-CDS2-R1 | TCACGGAAGGAAGAGGTCTTG |
| OsWRKY8-CDS2-F2 | TATCAAGCTAATTCGAGCTCATGTCTGGACCTGGAGGAGGAG |
| OsWRKY8-CDS2-R2 | CTAGAGGATCCCCGGGTACCTCACGGAAGGAAGAGGTCTTGC |
| OsWRKY10-CDS-F1 | ATGGCGGCTTCGCTGGGAC |
| OsWRKY10-CDS-R1 | TCAGAACGACGATTCCGACGAGT |
| OsWRKY10-CDS-F2 | TATCAAGCTAATTCGAGCTCATGGCGGCTTCGCTGGGAC |
| OsWRKY10-CDS-R2 | CTAGAGGATCCCCGGGTACCTCAGAACGACGATTCCGACGAG |
| OsWRKY28-CDS-F1 | ATGACCGCCGCGCCGGGG |
| OsWRKY28-CDS-R1 | TCAGTTCTTGGTCGGCGA |
| OsWRKY28-CDS-F2 | GCTAATTCGAGCTCGGTACCATGACCGCCGCGCCGGGG |
| OsWRKY28-CDS-R2 | CGACTCTAGAGGATCCTCAGTTCTTGGTCGGCGA |
| OsWRKY72-CDS-F1 | ATGGAGAACTTCCCGATACTCTT |
| OsWRKY72-CDS-R1 | CTACTGGAACATGTGGGAAGC |
| OsWRKY72-CDS-F2 | TATCAAGCTAATTCGAGCTCATGGAGAACTTCCCGATACTC |
| OsWRKY72-CDS-R2 | CTAGAGGATCCCCGGGTACCCTACTGGAACATGTGGGAAGC |
| OsWRKY111-CDS-F1 | ATGACCATCACACACGCTAG |
| OsWRKY111-CDS-R1 | CTAGAGCGACAGCCGGCGTCCA |
| OsWRKY111-CDS-F2 | TATCAAGCTAATTCGAGCTCATGACCATCACACACGCTAGC |
| OsWRKY111-CDS-R2 | CTAGAGGATCCCCGGGTACCCTAGAGCGACAGCCGGCG |
| OsWRKY6-CDS-F1 | ATGGATGGGGTGGAGGAG |
| OsWRKY6-CDS-R1 | TCAGGTCTGGGGATTAGCGG |
| OsWRKY6-CDS-F2 | TATCAAGCTAATTCGAGCTCATGGATGGGGTGGAGGAG |
| OsWRKY6-CDS-R2 | CTAGAGGATCCCCGGGTACCTCAGGTCTGGGGATTAGCGG |
| OsWRKY45-CDS-F1 | ATGACGTCATCGATGTCGC |
| OsWRKY45-CDS-R1 | TCAAAAGCTCAAACCCATAATGTCGT |
| OsWRKY45-CDS-F2 | TATCAAGCTAATTCGAGCTCATGACGTCATCGATGTCGC |
| OsWRKY45-CDS-R2 | CTAGAGGATCCCCGGGTACCTCAAAAGCTCAAACCCATAATGT |
| OsWRKY51-CDS-F1 | ATGATTACCATGGATCTGATGGGTGGG |
| OsWRKY51-CDS-R1 | TCAGGCGAGCGGCAGCGCCAG |
| OsWRKY51-CDS-F2 | TATCAAGCTAATTCGAGCTCATGATTACCATGGATCTGATGGGTGGG |
| OsWRKY51-CDS-R2 | CTAGAGGATCCCCGGGTACCTCAGGCGAGCGGCAGCGCCAG |
| OsPR10a-RT-F | GGGCACCATCTACACCATGAA |
| OsPR10a-RT-R | TCGTACTCCACCTTGAGCTT |
| OsAct1-RT-F | GAGTATGATGAGTCGGGTCCAG |
| OsAct1-RT-R | ACACCAACAATCCCAAACAGAG |
| [1] | Agrawal G K, Rakwal R, Jwa N S. 2000. Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochem Biophy Res Commun, 278(2): 290-298. |
| [2] | Agarwal P, Agarwal P K. 2014. Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Mol Biol Rep, 41(2): 599-611. |
| [3] | Andrade L B S, Oliveira A S, Ribeiro J K C, Kiyota S, Vasconcelos I M, de Oliveira J T, de Sales M P. 2010. Effects of a novel pathogenesis-related class 10 (PR-10) protein from Crotalaria pallida roots with papain inhibitory activity against root-knot nematode Meloidogyne incognita. J Agric Food Chem, 58(7): 4145-4152. |
| [4] | Brederode F T, Linthorst H J M, Bol J F. 1991. Differential induction of acquired resistance and PR gene expression in tobacco by virus infection, ethephon treatment, UV light and wounding. Plant Mol Biol, 17(6): 1117-1125. |
| [5] | Breiteneder H, Pettenburger K, Bito A, Valenta R, Kraft D, Rumpold H, Scheiner O, Breitenbach M. 1989. The gene coding for the major birch pollen allergen Betv1, is highly homologous to a pea disease resistance response gene. EMBO J, 8(7): 1935‒1938. |
| [6] | Breiteneder H, Hoffmann-Sommergruber K, O’Riordain G, Susani M, Ahorn H, Ebner C, Kraft D, Scheiner O. 1995. Molecular characterization of Api g 1, the major allergen of celery (Apium graveolens), and its immunological and structural relationships to a group of 17-kDa tree pollen allergens. Eur J Biochem, 233(2): 484-489. |
| [7] | Brown R L, Kazan K, McGrath K C, Maclean D J, Manners J M. 2003. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol, 132(2): 1020-1032. |
| [8] | Cai Q, Yuan Z, Chen M J, Yin C S, Luo Z J, Zhao X X, Liang W Q, Hu J P, Zhang D B. 2014. Jasmonic acid regulates spikelet development in rice. Nat Commun, 5: 3476. |
| [9] | Chen S B, Tao L Z, Zeng L R, Vega-Sanchez M E, Umemura K, Wang G L. 2006. A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol, 7(5): 417-427. |
| [10] | Choi C, Hwang S H, Fang I R, Kwon S I, Park S R, Ahn I, Kim J B, Hwang D J. 2015. Molecular characterization of Oryza sativa WRKY6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol, 208(3): 846-859. |
| [11] | Choi J, Huh S U, Kojima M, Sakakibara H, Paek K H, Hwang I. 2010. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell, 19(2): 284-295. |
| [12] | Dhatterwal P, Basu S, Mehrotra S, Mehrotra R. 2019. Genome wide analysis of W-box element in Arabidopsis thaliana reveals TGAC motif with genes down regulated by heat and salinity. Sci Rep, 9(1): 1681. |
| [13] | Dou S J, Guan M L, Li L Y, Liu G Z. 2014. Pathogenesis-related genes in rice. Chin Sci Bull, 59: 245‒258. (in Chinese with English abstract) |
| [14] | Durrant W E, Dong X. 2004. Systemic acquired resistance. Annu Rev Phytopathol, 42: 185-209. |
| [15] | Farmer E E, Almeras E, Krishnamurthy V. 2003. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr Opin Plant Biol, 6(4): 372-378. |
| [16] | Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico J M, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla J M, Pauwels L, Witters E, Puga M I, Paz-Ares J, Goossens A, Reymond P, de Jaeger G, Solano R. 2011. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell, 23(2): 701-715. |
| [17] | Feys B J, Parker J E. 2000. Interplay of signaling pathways in plant disease resistance. Trends Genet, 16(10): 449-455. |
| [18] | Fitzgerald H A, Canlas P E, Chern M S, Ronald P C. 2005. Alteration of TGA factor activity in rice results in enhanced tolerance to Xanthomonas oryzae pv. oryzae. Plant J, 43(3): 335-347. |
| [19] | Gutierrez-Alcala G, Calo L, Gros F, Caissard J C, Gotor C, Romero L C. 2005. A versatile promoter for the expression of proteins in glandular and non-glandular trichomes from a variety of plants. J Exp Bot, 56: 2487-2494. |
| [20] | Hashimoto M, Kisseleva L, Sawa S, Furukawa T, Komatsu S, Koshiba T. 2004. A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol, 45(5): 550-559. |
| [21] | Heyl A, Schmulling T. 2003. Cytokinin signal perception and transduction. Curr Opin Plant Biol, 6(5): 480-488. |
| [22] | Huang L F, Lin K H, He S L, Chen J L, Jiang J Z, Chen B H, Hou Y S, Chen R S, Hong C Y, Ho S L. 2016. Multiple patterns of regulation and overexpression of a ribonuclease-like pathogenesis- related protein gene,OsPR10a, conferring disease resistance in rice and Arabidopsis. PLoS One, 11(6): e0156414. |
| [23] | Hwang S H, Lee I A, Yie S W, Hwang D J. 2008. Identification of an OsPR10a promoter region responsive to salicylic acid. Planta, 227(5): 1141-1150. |
| [24] | Hwang S H, Kwon S I, Jang J Y, Fang I L, Lee H, Choi C, Park S, Ahn I, Bae S C, Hwang D J. 2016. OsWRKY51, a rice transcription factor, functions as a positive regulator in defense response against Xanthomonas oryzae pv. oryzae. Plant Cell Rep, 35(9): 1975‒1985. |
| [25] | Ito Y, Kurata N. 2006. Identification and characterization of cytokinin-signalling gene families in rice. Gene, 382: 57-65. |
| [26] | Iwai T, Seo S, Mitsuhara I, Ohashi Y. 2007. Probenazole-induced accumulation of salicylic acid confers resistance to Magnaporthe grisea in adult rice plants. Plant Cell Physiol, 48(7): 915-924. |
| [27] | Jameson P E. 2000. Cytokinins and auxins in plant-pathogen interactions: An overview. Plant Growth Regul, 32(2): 369-380. |
| [28] | Jefferson R A, Kavanagh T A, Bevan M W. 1987. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J, 6(13): 3901-3907. |
| [29] | Jia Y L, Valent B, Lee F N. 2003. Determination of host responses to Magnaporthe grisea on detached rice leaves using a spot inoculation method. Plant Dis, 87(2): 129-133. |
| [30] | Jiang C J, Shimono M, Sugano S, Kojima M, Liu X Q, Inoue H, Sakakibara H, Takatsuji H. 2013. Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Mol Plant Microbe Interact, 26(3): 287-296. |
| [31] | Jwa N S, Kumar Agrawal G, Rakwal R, Park C H, Prasad Agrawal V. 2001. Molecular cloning and characterization of a novel jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem Biophys Res Commun, 286(5): 973-983. |
| [32] | Jwa N S, Agrawal G K, Tamogami S, Yonekura M, Han O, Iwahashi H, Rakwal R. 2006. Role of defense/stress-related marker genes, proteins and secondary metabolites in defining rice self-defense mechanisms. Plant Physiol Biochem, 44: 261-273. |
| [33] | Kauffman H E, Reddy A P K, Hsieh S P Y, Merca S D. 1973. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep, 57(6): 537-541. |
| [34] | Kim S G, Kim S T, Wang Y M, Yu S, Choi I S, Kim Y C, Kim W T, Agrawal G K, Rakwal R, Kang K Y. 2011. The RNase activity of rice probenazole-induced protein1 (PBZ1) plays a key role in cell death in plants. Mol Cells, 31(1): 25-31. |
| [35] | Kim S T, Cho K S, Yu S, Kim S G, Hong J C, Han C D, Bae D W, Nam M H, Kang K Y. 2003. Proteomic analysis of differentially expressed proteins induced by rice blast fungus and elicitor in suspension-cultured rice cells. Proteomics, 3(12): 2368-2378. |
| [36] | Kim S T, Kim S G, Hwang D H, Kang S Y, Kim H J, Lee B H, Lee J J, Kang K Y. 2004. Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics, 4(11): 3569-3578. |
| [37] | Kim S T, Kim S G, Kang Y H, Wang Y M, Kim J Y, Yi N, Kim J K, Rakwal R, Koh H J, Kang K Y. 2008. Proteomics analysis of rice lesion mimic mutant (spl1) reveals tightly localized probenazole- induced protein (PBZ1) in cells undergoing programmed cell death. J Proteome Res, 7(4): 1750-1760. |
| [38] | Lee M W, Qi M, Yang Y. 2001. A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol Plant Microbe Interact, 14(4): 527-535. |
| [39] | Liu J J, Ekramoddoullah A K M. 2006. The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiol Mol Plant Pathol, 68(1): 3-13. |
| [40] | Lorenzo O, Chico J M, Sanchez-Serrano J J, Solano R. 2004. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell, 16(7): 1938‒1950. |
| [41] | Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton K A, Dangl J L, Dietrich R A. 2000. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet, 26(4): 403-410. |
| [42] | McGee J D, Hamer J E, Hodges T K. 2001. Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Mol Plant Microbe Interact, 14(7): 877-886. |
| [43] | Midoh N, Iwata M. 1996. Cloning and characterization of a probenazole-inducible gene for an intracellular pathogenesis- related protein in rice. Plant Cell Physiol, 37(1): 9-18. |
| [44] | Mitsuhara I, Iwai T, Seo S, Yanagawa Y, Kawahigasi H, Hirose S, Ohkawa Y, Ohashi Y. 2008. Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol Genet Genom, 279(4): 415-427. |
| [45] | Moons A, Prinsen E, Bauw G, Van Montagu M. 1997. Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell, 9(12): 2243-2259. |
| [46] | Park C J, Kim K J, Shin R, Park J M, Shin Y C, Paek K H. 2004. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J, 37(2): 186-198. |
| [47] | Peng Y, Bartley L E, Chen X W, Dardick C, Chern M, Ruan R, Canlas P E, Ronald P C. 2008. OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant, 1(3): 446-458. |
| [48] | Pfaffl M W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res, 29(9): e45. |
| [49] | Raines T, Blakley I C, Tsai Y C, Worthen J M, Franco-Zorrilla J M, Solano R, Schaller G E, Loraine A E, Kieber J J. 2016. Characterization of the cytokinin-responsive transcriptome in rice. BMC Plant Biol, 16(1): 260. |
| [50] | Rakwal R, Agrawal G K, Yonekura M. 2001a. Light-dependent induction of OsPR10 in rice(Oryza sativa L.) seedlings by the global stress signaling molecule jasmonic acid and protein phosphatase 2A inhibitors. Plant Sci, 161(3): 469-479. |
| [51] | Rakwal R, Agrawal G K, Agrawal V P. 2001b. Jasmonate, salicylate, protein phosphatase 2A inhibitors and kinetin up-regulate OsPR5 expression in cut-responsive rice(Oryza sativa). J Plant Physiol, 158(10): 1357-1362. |
| [52] | Reymond P, Farmer E E. 1998. Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol, 1(5): 404-411. |
| [53] | Ryu H S, Han M, Lee S K, Cho J I, Ryoo N, Heu S, Lee Y H, Bhoo S H, Wang G L, Hahn T R, Jeon J S. 2006. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep, 25(8): 836-847. |
| [54] | Sakai H, Aoyama T, Oka A. 2000. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J, 24(6): 703-711. |
| [55] | Schindler U, Beckmann H, Cashmore A R. 1992. TGA1 and G-box binding factors: Two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell, 4(10): 1309-1319. |
| [56] | Schweizer F, Fernandez-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, Lewsey M G, Ecker J R, Solano R, Reymond P. 2013. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell, 25(8): 3117-3132. |
| [57] | Sels J, Mathys J, De Coninck B M A, Cammue B P A, De Bolle M F C. 2008. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol Biochem, 46(11): 941-950. |
| [58] | Serna L, Martin C. 2006. Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci, 11(6): 274-280. |
| [59] | Shimono M, Sugano S, Nakayama A, Jiang C J, Ono K, Toki S, Takatsuji H. 2007. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell, 19(6): 2064‒2076. |
| [60] | Takeuchi K, Gyohda A, Tominaga M, Kawakatsu M, Hatakeyama A, Ishii N, Shimaya K, Nishimura T, Riemann M, Nick P, Hashimoto M, Komano T, Endo A, Okamoto T, Jikumaru Y, Kamiya Y, Terakawa T, Koshiba T. 2011. RSOsPR10 expression in response to environmental stresses is regulated antagonistically by jasmonate/ethylene and salicylic acid signaling pathways in rice roots. Plant Cell Physiol, 52(9): 1686-1696. |
| [61] | Talbot N J, Ebbole D J, Hamer J E. 1993. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell, 5(11): 1575-1590. |
| [62] | Thaler J S, Humphrey P T, Whiteman N K. 2012. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci, 17(5): 260-270. |
| [63] | Tsai Y C, Weir N R, Hill K, Zhang W J, Kim H J, Shiu S H, Schaller G E, Kieber J J. 2012. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol, 158(4): 1666-1684. |
| [64] | van Loon L C. 1985. Pathogenesis-related proteins. Plant Mol Biol, 4: 111-116. |
| [65] | van Loon L C, Pierpoint W S, Boller T, Conejero V. 1994. Recommendations for naming plant pathogenesis-related proteins. Plant Mol Biol Rep, 12(3): 245-264. |
| [66] | van Loon L C, van Strien E A. 1999. The families of pathogenesis- related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol, 55(2): 85-97. |
| [67] | van Loon L C, Rep M, Pieterse C M. 2006. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopath, 44: 135-162. |
| [68] | Wu Q, Hou M M, Li L Y, Liu L J, Hou Y X, Liu G Z. 2011. Induction of pathogenesis-related proteins in rice bacterial blight resistant gene Xa21-mediated interactions with Xanthomnas oryzae pv. oryzae. J Plant Pathol, 93(2): 455-459. |
| [69] | Xie Y R, Chen Z Y, Brown R L, Bhatnagar D. 2010. Expression and functional characterization of two pathogenesis-related protein 10 genes from Zea mays. J Plant Physiol, 167(2): 121-130. |
| [70] | Xiong M, Meng S, Qiu J H, Shi H B, Shen X L, Kou Y J. 2020. Putative phosphatase UvPsr1 is required for mycelial growth, conidiation, stress response and pathogenicity in Ustilaginonidea virens. Rice Sci, 20(6): 529-536. |
| [71] | Xu Z M, Li L H, Gao X Q, Yuan Z J, Li Xin, Tian X D, Wang L L, Qu S H. 2020. Comparative transcriptome analysis of transgenic rice line carrying the rice blast resistance gene Pi9. Chin J Rice Sci, 34(3): 245‒255. (in Chinese with English abstract) |
| [72] | Yamamoto T, Yoshida Y, Nakajima K, Tominaga M, Gyohda A, Suzuki H, Okamoto T, Nishimura T, Yokotani N, Minami E, Nishizawa Y, Miyamoto K, Yamane H, Okada K, Koshiba T. 2018. Expression of RSOsPR10 in rice roots is antagonistically regulated by jasmonate/ethylene and salicylic acid via the activator OsERF87 and the repressor OsWRKY76, respectively. Plant Direct, 2(3): e00049. |
| [73] | Yang B, Bogdanove A. 2013. Inoculation and virulence assay for bacterial blight and bacterial leaf streak of rice. In: Yang Y. Rice Protocols. Methods in Molecular Biology (Methods and Protocls). Vol 956. Totowa: Humana Press: 249‒255. |
| [74] | Yoshida S, Forno D A, Cock J H, Gomez K A. 1976. Laboratory Manual for Physiological Studies of Rice. Manila, the Philippines: International Rice Research Institute. |
| [75] | Yu F B, Zhou J, Wang X M, Yang Y, Yu C L, Cheng Y, Yan C Q, Chen J P. 2014. Expression characteristics of rice OsPR1b gene promoter. Lett Biotechnol, 25(4): 24‒30. (in Chinese with English abstract) |
| [76] | Zeevaart J A D, Creelman R A. 1988. Metabolism and physiology of abscisic acid. Ann Rev Plant Physiol Plant Mol Biol, 39(1): 439-473. |
| [77] | Zhang J, Peng Y L, Guo Z J. 2008. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res, 18(4): 508-521. |
| [78] | Zhang Y, Su J B, Duan S, Ao Y, Dai J R, Liu J, Wang P, Li Y G, Liu B, Feng D R, Wang J F, Wang H B. 2011. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods, 7(1): 30. |
| [79] | Zhou J, Yang Y, Wang X M, Yu F B, Yu C L, Chen J, Cheng Y, Yan C Q, Chen J P. 2013. Enhanced transgene expression in rice following selection controlled by weak promoters. BMC Biotechnol, 13: 29. |
| [80] | Zhou J, Yu F B, Wang X M, Yang Y, Yu C L, Liu H J, Cheng Y, Yan C Q, Chen J P. 2014. Specific expression of DR5 promoter in rice roots using a tCUP derived promoter-reporter system. PLoS One, 9(1): e87008. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [13] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||