
Rice Science ›› 2018, Vol. 25 ›› Issue (5): 293-296.DOI: 10.1016/j.rsci.2018.08.004
• Orginal Article • Previous Articles
Lee Jae-Sung1,2, Wissuwa Matthias3( ), B. Zamora Oscar1, M. Ismail Abdelbagi2(
), B. Zamora Oscar1, M. Ismail Abdelbagi2( )
)
Received:2018-03-14
Accepted:2018-05-30
Online:2018-09-28
Published:2018-06-11
Lee Jae-Sung, Wissuwa Matthias, B. Zamora Oscar, M. Ismail Abdelbagi. Novel Sources of aus Rice for Zinc Deficiency Tolerance Identified Through Association Analysis Using High-Density SNP Array[J]. Rice Science, 2018, 25(5): 293-296.
Add to citation manager EndNote|Ris|BibTeX
| Locus ID | Chr | Position | Annotation | Expression a | |||
|---|---|---|---|---|---|---|---|
| LOC_Os07g45090 | 7 | 26 913 464-26 917 496 | NADH-ubiquinone oxidoreductase | E (2.84 × 10-2), G (5.94 × 10-3) | |||
| LOC_Os07g45170 | 7 | 26 965 232-26 967 050 | Protein phosphatase | E (1.13 × 10-2) | |||
| LOC_Os09g26160 | 9 | 15 765 945-15 770 861 | Glutamate receptor | E (2.58 × 10-4), E × G (2.00 × 10-2) | |||
| LOC_Os09g26190 | 9 | 15 790 768-15 800 951 | CBS domain containing protein | E (2.10 × 10-2), G (2.63 × 10-2) | |||
| Locus ID | Network-ontology b | Significant marker position | P-value (R2, %) | Allele | |||
| LOC_Os07g45090 | Oxidation reduction; response to zinc ion and salt stress | na | na | na | |||
| LOC_Os07g45170 | Response to oxidative and salt stresses; double-strand break repair | na | na | na | |||
| LOC_Os09g26160 | Response to salt stress; ascorbic acid biosynthesis | 15 770 497 (4th exon) | 2.10 × 10-5 (24) | A/G | |||
| LOC_Os09g26190 | Photosynthesis; chlorophyll biosynthesis; protein to chloroplast thylakoid membrane | 15 787 904 (3 kb upstream of 5′) | 6.27 × 10-6 (28) | G/C | |||
Table 1 List of a priori candidate genes associated with Zn deficiency tolerance.
| Locus ID | Chr | Position | Annotation | Expression a | |||
|---|---|---|---|---|---|---|---|
| LOC_Os07g45090 | 7 | 26 913 464-26 917 496 | NADH-ubiquinone oxidoreductase | E (2.84 × 10-2), G (5.94 × 10-3) | |||
| LOC_Os07g45170 | 7 | 26 965 232-26 967 050 | Protein phosphatase | E (1.13 × 10-2) | |||
| LOC_Os09g26160 | 9 | 15 765 945-15 770 861 | Glutamate receptor | E (2.58 × 10-4), E × G (2.00 × 10-2) | |||
| LOC_Os09g26190 | 9 | 15 790 768-15 800 951 | CBS domain containing protein | E (2.10 × 10-2), G (2.63 × 10-2) | |||
| Locus ID | Network-ontology b | Significant marker position | P-value (R2, %) | Allele | |||
| LOC_Os07g45090 | Oxidation reduction; response to zinc ion and salt stress | na | na | na | |||
| LOC_Os07g45170 | Response to oxidative and salt stresses; double-strand break repair | na | na | na | |||
| LOC_Os09g26160 | Response to salt stress; ascorbic acid biosynthesis | 15 770 497 (4th exon) | 2.10 × 10-5 (24) | A/G | |||
| LOC_Os09g26190 | Photosynthesis; chlorophyll biosynthesis; protein to chloroplast thylakoid membrane | 15 787 904 (3 kb upstream of 5′) | 6.27 × 10-6 (28) | G/C | |||
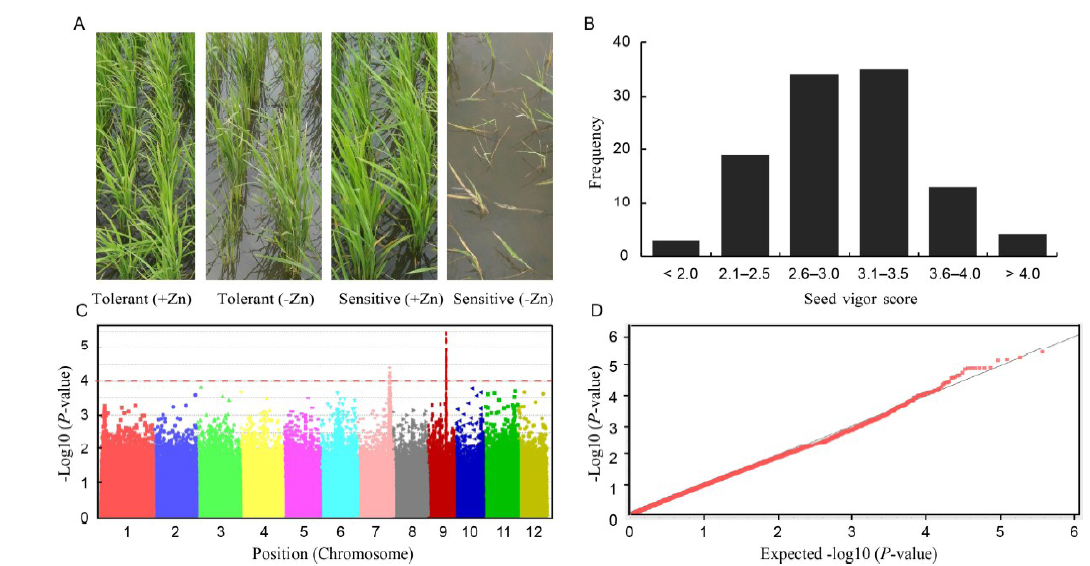
Fig. 1. Genome-wide association analysis of plant vigor of a diverse panel of aus accessions under Zn deficiency.Data was collected at fourth week after transplanting during the peak-stress period.A, Plant growth of tolerant and sensitive accessions under control (+Zn) and Zn deficiency (-Zn). B, Distribution of seed vigor scores. Higher scores indicate higher tolerance to Zn deficiency. C, Manhattan plot. Plots above the threshold line indicate significant association. D, Quantile-quantile plot.
| 1 | Alloway B J.2008. Zinc in Soils and Crop Nutrition. 2nd edition. International Zinc Association and International Fertilizer Industry Association. Brussels, Belgium and Paris, France. |
| 2 | Cakmak I.2000. Possible roles of zinc in protecting plant cells from damage by reactive oxygen species.New Phytol, 146(2): 185-205. |
| 3 | Famoso A N, Zhao K, Clark R T, Tung C W, Wright M H, Bustamante C, Kochian L V, McCouch S R.2011. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping.PLoS Genet, 7(8): e1002221. |
| 4 | Frei M, Wang Y X, Ismail A M, Wissuwa M.2010. Biochemical factors conferring shoot tolerance to oxidative stress in rice grown in low zinc soil.Funct Plant Biol, 37(1): 74-84. |
| 5 | Impa S M, Johnson-Beebout S E.2012. Mitigating zinc deficiency and achieving high grain Zn in rice through integration of soil chemistry and plant physiology research.Plant Soil, 361: 3-41. |
| 6 | Khush G S.1997. Origin, dispersal, cultivation and variation of rice.Plant Mol Biol, 35: 25-34. |
| 7 | Lee J S, Sajise A G C, Gregorio G B, Kretzschmar T, Ismail A M, Wissuwa M.2017a. Genetic dissection for zinc deficiency tolerance in rice using bi-parental mapping and association analysis.Theor Appl Genet, 130(9): 1903-1914. |
| 8 | Lee J S, Wissuwa M, Zamora O B, Ismail A M.2017b. Biochemical indicators of root damage in rice (Oryza sativa L.) genotypes under zinc deficiency stress.J Plant Res, 30(6): 1071-1077. |
| 9 | Londo J P, Chiang Y C, Hung K H, Chiang T Y, Schaal B A.2006. Phylogeography of Asian wild rice,Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa.Proc Natl Acad Sci USA, 103(25): 9578-9583. |
| 10 | McCouch S R, Wright M H, Tung C W, Maron L G, McNally K L, Fitzgerald M, Singh N, DeClerck G, Agosto-Perez F, Korniliev P, Greenberg A J, Naredo M E B, Mercado S M Q, Harrington S E, Shi Y, Branchini D A, Kuser-Falcaõ P R, Leung H, Ebana K, Yano M, Eizenga G, McClung A, Mezey J.2016. Open access resources for genome-wide association mapping in rice.Nat Commun, 7: 10532. |
| 11 | Neue H U, Lantin R S.1994. Micronutrient toxicities and deficiencies in rice. In: Yeo A R, Flowers T J. Soil Mineral Stresses: Approaches to Crop Improvement. Berlin: Springer- Verlag: 175-200. |
| 12 | Quijano-Guerta C, Kirk G J D, Portugal A M, Bartolome V I, McLaren G C.2002. Tolerance of rice germplasm to zinc deficiency.Field Crop Res, 76: 123-130. |
| 13 | Singh B, Natesan S K A, Singh B K, Usha K.2003. Improving zinc efficiency of cereals under zinc deficiency.Curr Sci, 88(1): 36-44. |
| 14 | Widodo X A, Broadley M R, Rose T, Frei M, Pariasca-Tanaka J, Yoshihashi T, Thomson M, Hammond J P, Aprile A, Close T J, Ismail A M, Wissuwa M.2010. Response to zinc deficiency of two rice lines with contrasting tolerance is determined by root growth maintenance and organic acid exudation rates, and not by zinc-transporter activity.New Phytol, 186(2): 400-414. |
| 15 | Wissuwa M, Ismail A M, Yanagihara S.2006. Effects of zinc deficiency on rice growth and genetic factors contributing to tolerance.Plant Physiol, 142(2): 731-741. |
| 16 | Yoshida S, Tanaka A.1969. Zinc deficiency of the rice plant in calcareous soils.Soil Sci Plant Nutr, 15(2): 75-80. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [13] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||