
Rice Science ›› 2022, Vol. 29 ›› Issue (1): 67-75.DOI: 10.1016/j.rsci.2021.12.006
• Research Paper • Previous Articles Next Articles
Wang Rui1,2,#, Zhang Dandan2,#, Li Shengnan2,#, Gao Jinlan2, Han Liebao1( ), Qiu Jinlong2,3(
), Qiu Jinlong2,3( )
)
Received:2020-12-17
Accepted:2021-03-01
Online:2022-01-28
Published:2022-01-01
Contact:
Han Liebao, Qiu Jinlong
About author:First author contact:#These authors contributed equally to this work
Wang Rui, Zhang Dandan, Li Shengnan, Gao Jinlan, Han Liebao, Qiu Jinlong. Simple Bioassay for PAMP-Triggered Immunity in Rice Seedlings Based on Lateral Root Growth Inhibition[J]. Rice Science, 2022, 29(1): 67-75.
Add to citation manager EndNote|Ris|BibTeX
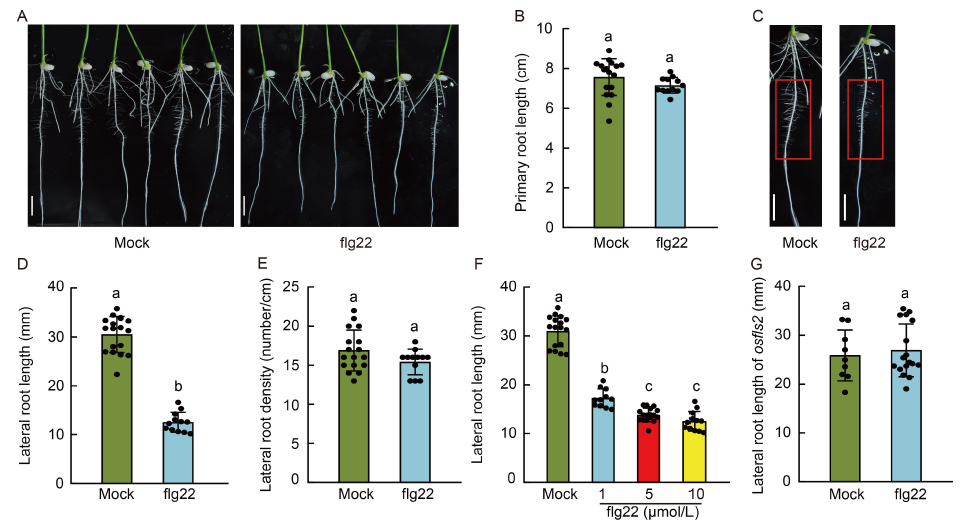
Fig. 1. Pathogen-associated molecular pattern (PAMP) flg22 inhibits lateral root growth of rice seedlings. A, Primary root growth following flg22 treatment. Oryza sativa ssp. japonica cultivar Nipponbare seedlings with a primary root length of 1.5?2.0 cm were transferred to half-strength Murashige and Skoog (MS) medium supplied with 10 μmol/L flg22 or without (mock) and grown vertically for 3 d before the data were collected. Scale bars, 1 cm. B, Primary root length of rice seedlings treated without (mock) or with 10 μmol/L flg22. C, Lateral root growth of rice seedlings treated without (mock) or with 10 μmol/L flg22. The primary root (1.5-4.5 cm) used for analyzing lateral root length and density is outlined in red. Scale bars, 1 cm. D and E, Lateral root length (D) and density (E) of rice seedlings treated without (mock) or with 10 μmol/L flg22. F, Effect of flg22 on lateral root growth inhibition is dose dependence. Lateral root length of rice seedlings was measured 3 d after grown vertically on half-strength MS medium with indicated flg22 concentrations. G, Lateral root length of the osfls2 mutant seedlings treated without (mock) or with 10 μmol/L flg22. Bars indicate Mean ± SD of n > 8 seedlings per treatment. The significance of the differences was determined by a one-way ANOVA with the Tukey’s multiple comparisons test. Different lowercase letters above the bars indicate significant differences at P < 0.01.
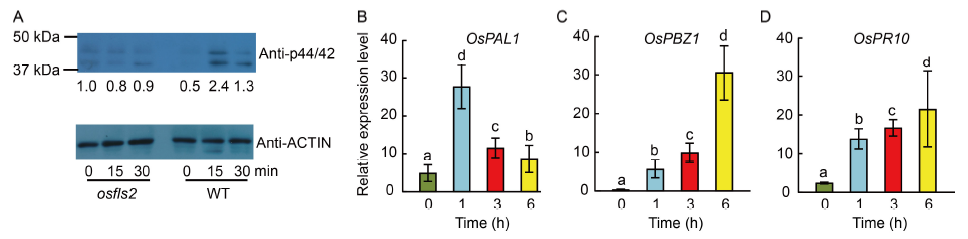
Fig. 2. flg22-induced responses in lateral roots of rice seedlings. A, Activation of mitogen-activated protein kinases (MAPKs) upon the 5 μmol/L flg22 treatment. MAPK activity was analyzed by immunoblotting with Phospho-p44/42 MAPK (Erk1/2) antibody (top panel), and ACTIN was used as a protein loading control (bottom panel). The intensity of the osfls2 mutant before the flg22 treatment was identified as ‘1.0’. WT, Wild type. B?D, Defense gene expression induced upon the flg22 treatment. The relative expression levels of OsPAL1 (B), OsPBZ1 (C) and OsPR10 (D) in the roots of rice seedlings treated with 5 μmol/L flg22 were determined by quantitative real-time PCR. OsActin2 was used as an internal reference. Data are presented as Mean ± SD of three biological replicates. Different lowercase letters above the bars indicate significant differences at P < 0.01.
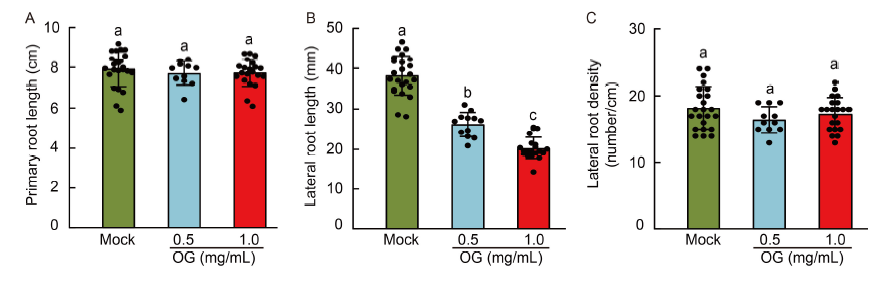
Fig. 3. Oligogalacturonides (OGs) inhibit lateral root growth of rice seedlings. A , Primary root length of rice seedlings following the OG treatment. B, Lateral root length of rice seedlings after the OG treatment. C, Lateral root density of rice seedlings after the OG treatment. Bars indicate Mean ± SD of n > 8 seedlings per treatment. The significance of the differences was determined by a one-way ANOVA with the Tukey’s multiple comparisons test. Different lowercase letters above the bars indicate significant differences at P < 0.01.
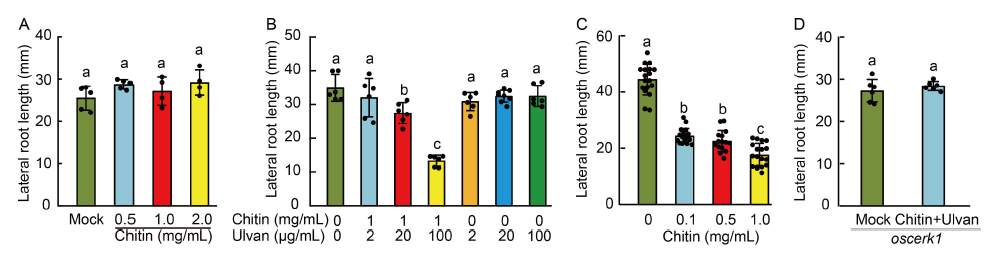
Fig. 4. Ulvan priming of rice seedlings is required for inhibition of lateral root growth by chitin. A , Lateral root length of rice seedlings following the chitin treatment. B, Lateral root length of rice seedlings primed with ulvan and treated with chitin. C, Lateral root length of rice seedlings treated with indicated concentrations of chitin and primed with 100 μg/mL ulvan. D, Lateral root length of oscerk1 mutant seedlings treated without (mock) or with 0.5 mg/mL chitin and 100 μg/mL ulvan. Bars indicate Mean ± SD of n ≥ 4 seedlings per treatment. The significance of the differences was determined by a one-way ANOVA with the Tukey’s multiple comparison test. Different lowercase letters above the bars indicate significant differences at P < 0.01.
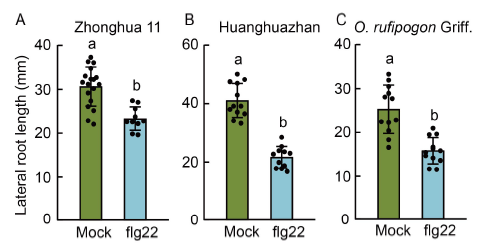
Fig. 5. Pathogen-associated molecular pattern (PAMP)-induced lateral root growth inhibition in various rice cultivars and wild species. A , Lateral root length of japonica rice Zhonghua 11 seedlings treated without (mock) or with 5 μmol/L flg22. B, Lateral root length of indica rice Huanghuazhan seedlings treated without (mock) or with 5 μmol/L flg22. C, Lateral root length of O. rufipogon Griff. seedlings treated without (mock) or with 5 μmol/L flg22. Bars indicate Mean ± SD of n > 8 seedlings per treatment. The significance of the differences was determined by a one-way ANOVA with the Tukey’s multiple comparison test. Different lowercase letters above the bars indicate significant differences at P < 0.01.
| [1] |
Akamatsu A, Wong H L, Fujiwara M, Okuda J, Nishide K, Uno K, Imai K, Umemura K, Kawasaki T, Kawano Y, Shimamoto K. 2013. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe, 13(4): 465-476.
PMID |
| [2] | Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez- Ibanez S, Chinchilla D, Rathjen J P, de Vries S C, Zipfel C. 2012. Brassinosteroids inhibit pathogen-associated molecular pattern- triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA, 109(1): 303-308. |
| [3] | Anderson J C, Bartels S, González Besteiro M A, Shahollari B, Ulm R, Peck S C. 2011. Arabidopsis MAP kinase phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J, 67(2): 258-268. |
| [4] | Ao Y, Li Z Q, Feng D R, Xiong F, Liu J, Li J F, Wang M L, Wang J F, Liu B, Wang H B. 2014. OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J, 80(6): 1072-1084. |
| [5] | Ausubel F M. 2005. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol, 6(10): 973-979. |
| [6] | Bajaj S, Mohanty A. 2005. Recent advances in rice biotechnology- towards genetically superior transgenic rice. Plant Biotechnol J, 3(3): 275-307. |
| [7] | Boller T, Felix G. 2009. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol, 60(1): 379-406. |
| [8] | Brutus A, Sicilia F, Macone A, Cervone F, de Lorenzo G. 2010. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA, 107(20): 9452-9457. |
| [9] | Cao Y R, Liang Y, Tanaka K, Nguyen C T, Jedrzejczak R, Joachimiak A, Stacey G. 2014. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife, 3(2): e03766. |
| [10] | Chen X W, Zuo S M, Schwessinger B, Chern M, Canlas P E, Ruan D L, Zhou X G, Wang J, Daudi A, Petzold C J, Heazlewood J L, Ronald P C. 2014. An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol Plant, 7(5): 874-892. |
| [11] |
Chuberre C, Plancot B, Driouich A, Moore J P, Bardor M, Gügi B, Vicré M. 2018. Plant immunity is compartmentalized and specialized in roots. Front Plant Sci, 9: 1692.
PMID |
| [12] | Conrath U, Beckers G J M, Langenbach C J G, Jaskiewicz M R. 2015. Priming for enhanced defense. Annu Rev Phytopathol, 53(1): 97-119. |
| [13] | Ding B, del Rosario Bellizzi M, Ning Y, Meyers B C, Wang L. 2012. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell, 24(9): 3783-3794. |
| [14] |
Ferrari S, Savatin D V, Sicilia F, Gramegna G, Cervone F, De Lorenzo G. 2013. Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci, 4: 49.
PMID |
| [15] |
Gómez-Gómez L, Boller T. 2000. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell, 5(6): 1003-1011.
PMID |
| [16] |
Gómez-Gómez L, Felix G, Boller T. 1999. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J, 18(3): 277-284.
PMID |
| [17] | Hann D R, Rathjen J P. 2007. Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J, 49(4): 607-618. |
| [18] | Howard R J. 1997. Breaching the outer barriers: Cuticle and cell wall penetration. In: Carroll G C, Tudzynski P. The Mycota: Plant Relationships, Part A. Springer-Verlag Berlin Heidelberg: 43-60. |
| [19] | Huang P Y, Yeh Y H, Liu A C, Cheng C P, Zimmerli L. 2014. The Arabidopsis LecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern- triggered immunity. Plant J, 79(2): 243-255. |
| [20] | Jiang L Y, Anderson J C, Besteiro M A G, Peck S C. 2017. Phosphorylation of Arabidopsis MAP kinase phosphatase 1 (MKP1) is required for PAMP responses and resistance against bacteria. Plant Physiol, 175(4): 1839-1852. |
| [21] | Jones J D G, Dangl J L. 2006. The plant immune system. Nature, 444: 323-329. |
| [22] | Laluk K, Luo H L, Chai M F, Dhawan R, Lai Z B, Mengiste T. 2011. Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell, 23(8): 2831-2849. |
| [23] | Li Y, Zhang Q Q, Zhang J G, Wu L, Qi Y J, Zhou J M. 2010. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol, 152(4): 2222-2231. |
| [24] |
Li Y, Zhang Y, Wang Q X, Wang T T, Cao X L, Zhao Z X, Zhao S L, Xu Y J, Xiao Z Y, Li J L, Fan J, Yang H, Huang F, Xiao S Y, Wang W M. 2018. RESISTANCE TO POWDERY MILDEW 8.1 boosts pattern-triggered immunity against multiple pathogens in Arabidopsis and rice. Plant Biotechnol J, 16(2): 428-441.
PMID |
| [25] | Liu H J, Wang S F, Yu X B, Yu J, He X W, Zhang S L, Shou H X, Wu P. 2005. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J, 43(1): 47-56. |
| [26] | Liu J L, Park C H, He F, Nagano M, Wang M, Bellizzi M, Zhang K, Zeng X S, Liu W D, Ning Y, Kawano Y, Wang G L. 2015. The RhoGAP SPIN6 associates with SPL11 and OsRac1 and negatively regulates programmed cell death and innate immunity in rice. PLoS Pathog, 11(2): e1004629. |
| [27] | Liu T T, Liu Z X, Song C J, Hu Y F, Han Z F, She J, Fan F F, Wang J W, Jin C W, Chang J B, Zhou J M, Chai J J. 2012. Chitin- induced dimerization activates a plant immune receptor. Science, 336: 1160-1164. |
| [28] | Nekrasov V, Li J, Batoux M, Roux M, Chu Z H, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, van Esse H P, Jorda L, Schwessinger B, Nicaise V, Thomma B P H J, Molina A, Jones J D G, Zipfel C. 2009. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J, 28(21): 3428-3438. |
| [29] | Paulert R, Ebbinghaus D, Urlass C, Moerschbacher B M. 2010. Priming of the oxidative burst in rice and wheat cell cultures by ulvan, a polysaccharide from green macroalgae, and enhanced resistance against powdery mildew in wheat and barley plants. Plant Pathol, 59(4): 634-642. |
| [30] | Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D. 2011. Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage- associated molecular patterns. Plant J, 68(1): 100-113. |
| [31] | Rebouillat J, Dievart A, Verdeil J L, Escoute J, Giese G, Breitler J C, Gantet P, Espeout S, Guiderdoni E, Périn C. 2009. Molecular genetics of rice root development. Rice, 2(1): 15-34. |
| [32] |
Robatzek S, Bittel P, Chinchilla D, Köchner P, Felix G, Shiu S H, Boller T. 2007. Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol Biol, 64(5): 539-547.
PMID |
| [33] |
Roberts R, Liu A E, Wan L W, Geiger A M, Hind S R, Rosli H G, Martin G B. 2020. Molecular characterization of differences between the tomato immune receptors flagellin sensing 3 and flagellin sensing 2. Plant Physiol, 183: 1825-1837.
PMID |
| [34] | Robin A H K, Saha P S. 2015. Morphology of lateral roots of twelve rice cultivars of Bangladesh: Dimension increase and diameter reduction in progressive root branching at the vegetative stage. Plant Root, 9: 34-42. |
| [35] |
Sanabria N, Goring D, Nürnberger T, Dubery I. 2008. Self/nonself perception and recognition mechanisms in plants: A comparison of self-incompatibility and innate immunity. New Phytol, 178(3): 503-514.
PMID |
| [36] | Sasaki O, Yamazaki K, Kawata S. 1981. The relationship between the diameters and the structures of lateral roots in rice plants. Jpn J Crop Sci, 50(4): 476-480. |
| [37] | Shan Q W, Wang Y P, Li J, Gao C X. 2014. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc, 9(10): 2395-2410. |
| [38] | Shibuya N, Minami E. 2001. Oligosaccharide signalling for defence responses in plant. Physiol Mol Plant Pathol, 59: 223-233. |
| [39] | Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, Shibuya N. 2010. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J, 64(2): 204-214. |
| [40] | Stegmann M, Anderson R G, Ichimura K, Pecenkova T, Reuter P, Žárský V, McDowell J M, Shirasu K, Trujillo M. 2012. The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell, 24(11): 4703-4716. |
| [41] | Takai R, Kaneda T, Isogai A, Takayama S, Che F S. 2007. A new method of defense response analysis using a transient expression system in rice protoplasts. Biosci Biotechnol Biochem, 71(2): 590-593. |
| [42] | Takai R, Isogai A, Takayama S, Che F S. 2008. Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol Plant Microbe Interact, 21(12): 1635-1642. |
| [43] | Wang S Z, Sun Z, Wang H Q, Liu L J, Lu F, Yang J, Zhang M, Zhang S Y, Guo Z J, Bent A F, Sun W X. 2015. Rice OsFLS2- mediated perception of bacterial flagellins is evaded by Xanthomonas oryzae pvs. oryzae and oryzicola. Mol Plant, 8(7): 1024-1037. |
| [44] | Xing H L, Dong L, Wang Z P, Zhang H Y, Han C Y, Liu B, Wang X C, Chen Q J. 2014. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol, 14(1): 327. |
| [45] | Xu J, Hong J H. 2013. Root development. In: Zhang Q F, Wing R A. Genetics and Genomics of Rice. New York, USA: Springer-Verlag, : 297-316. |
| [46] | Yang C, Yu Y Q, Huang J K, Meng F W, Pang J H, Zhao Q Q, Islam M A, Xu N, Tian Y, Liu J. 2019. Binding of the Magnaporthe oryzae chitinase MoChia1 by a rice tetratricopeptide repeat protein allows free chitin to trigger immune responses. Plant Cell, 31(1): 172-188. |
| [47] |
Zhang J, Shao F, Li Y, Cui H T, Chen L J, Li H T, Zou Y, Long C Z, Lan L F, Chai J J, Chen S, Tang X Y, Zhou J M. 2007. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe, 1(3): 175-185.
PMID |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [13] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||